
Enzyme kinetics
Encyclopedia

Chemical reaction
A chemical reaction is a process that leads to the transformation of one set of chemical substances to another. Chemical reactions can be either spontaneous, requiring no input of energy, or non-spontaneous, typically following the input of some type of energy, such as heat, light or electricity...
s that are catalysed by enzymes. In enzyme kinetics, the reaction rate
Reaction rate
The reaction rate or speed of reaction for a reactant or product in a particular reaction is intuitively defined as how fast or slow a reaction takes place...
is measured and the effects of varying the conditions of the reaction investigated. Studying an enzyme's kinetics
Chemical kinetics
Chemical kinetics, also known as reaction kinetics, is the study of rates of chemical processes. Chemical kinetics includes investigations of how different experimental conditions can influence the speed of a chemical reaction and yield information about the reaction's mechanism and transition...
in this way can reveal the catalytic mechanism of this enzyme, its role in metabolism
Metabolism
Metabolism is the set of chemical reactions that happen in the cells of living organisms to sustain life. These processes allow organisms to grow and reproduce, maintain their structures, and respond to their environments. Metabolism is usually divided into two categories...
, how its activity is controlled, and how a drug
Drug
A drug, broadly speaking, is any substance that, when absorbed into the body of a living organism, alters normal bodily function. There is no single, precise definition, as there are different meanings in drug control law, government regulations, medicine, and colloquial usage.In pharmacology, a...
or an agonist might inhibit
Enzyme inhibitor
An enzyme inhibitor is a molecule that binds to enzymes and decreases their activity. Since blocking an enzyme's activity can kill a pathogen or correct a metabolic imbalance, many drugs are enzyme inhibitors. They are also used as herbicides and pesticides...
the enzyme.
Enzymes are usually protein
Protein
Proteins are biochemical compounds consisting of one or more polypeptides typically folded into a globular or fibrous form, facilitating a biological function. A polypeptide is a single linear polymer chain of amino acids bonded together by peptide bonds between the carboxyl and amino groups of...
molecule
Molecule
A molecule is an electrically neutral group of at least two atoms held together by covalent chemical bonds. Molecules are distinguished from ions by their electrical charge...
s that manipulate other molecules — the enzymes' substrate
Substrate (biochemistry)
In biochemistry, a substrate is a molecule upon which an enzyme acts. Enzymes catalyze chemical reactions involving the substrate. In the case of a single substrate, the substrate binds with the enzyme active site, and an enzyme-substrate complex is formed. The substrate is transformed into one or...
s. These target molecules bind to an enzyme's active site
Active site
In biology the active site is part of an enzyme where substrates bind and undergo a chemical reaction. The majority of enzymes are proteins but RNA enzymes called ribozymes also exist. The active site of an enzyme is usually found in a cleft or pocket that is lined by amino acid residues that...
and are transformed into products
Product (biology)
In biochemistry, a product is something "manufactured" by an enzyme from its substrate. For example, the products of lactase are galactose and glucose, which are produced from the substrate lactose....
through a series of steps known as the enzymatic mechanism
Enzyme catalysis
Enzyme catalysis is the catalysis of chemical reactions by specialized proteins known as enzymes. Catalysis of biochemical reactions in the cell is vital due to the very low reaction rates of the uncatalysed reactions....
. These mechanisms can be divided into single-substrate and multiple-substrate mechanisms. Kinetic studies on enzymes that only bind one substrate, such as triosephosphate isomerase
Triosephosphateisomerase
Triose-phosphate isomerase , is an enzyme that catalyzes the reversible interconversion of the triose phosphate isomers dihydroxyacetone phosphate and D-glyceraldehyde 3-phosphate....
, aim to measure the affinity
Dissociation constant
In chemistry, biochemistry, and pharmacology, a dissociation constant is a specific type of equilibrium constant that measures the propensity of a larger object to separate reversibly into smaller components, as when a complex falls apart into its component molecules, or when a salt splits up into...
with which the enzyme binds this substrate and the turnover rate.
When enzymes bind multiple substrates, such as dihydrofolate reductase
Dihydrofolate reductase
- Function :Dihydrofolate reductase converts dihydrofolate into tetrahydrofolate, a methyl group shuttle required for the de novo synthesis of purines, thymidylic acid, and certain amino acids...
(shown right), enzyme kinetics can also show the sequence in which these substrates bind and the sequence in which products are released. An example of enzymes that bind a single substrate and release multiple products are protease
Protease
A protease is any enzyme that conducts proteolysis, that is, begins protein catabolism by hydrolysis of the peptide bonds that link amino acids together in the polypeptide chain forming the protein....
s, which cleave one protein substrate into two polypeptide products. Others join two substrates together, such as DNA polymerase
DNA polymerase
A DNA polymerase is an enzyme that helps catalyze in the polymerization of deoxyribonucleotides into a DNA strand. DNA polymerases are best known for their feedback role in DNA replication, in which the polymerase "reads" an intact DNA strand as a template and uses it to synthesize the new strand....
linking a nucleotide
Nucleotide
Nucleotides are molecules that, when joined together, make up the structural units of RNA and DNA. In addition, nucleotides participate in cellular signaling , and are incorporated into important cofactors of enzymatic reactions...
to DNA
DNA
Deoxyribonucleic acid is a nucleic acid that contains the genetic instructions used in the development and functioning of all known living organisms . The DNA segments that carry this genetic information are called genes, but other DNA sequences have structural purposes, or are involved in...
. Although these mechanisms are often a complex series of steps, there is typically one rate-determining step that determines the overall kinetics. This rate-determining step
Rate-determining step
The rate-determining step is a chemistry term for the slowest step in a chemical reaction. The rate-determining step is often compared to the neck of a funnel; the rate at which water flows through the funnel is determined by the width of the neck, not by the speed at which water is poured in. In...
may be a chemical reaction or a conformational
Conformational isomerism
In chemistry, conformational isomerism is a form of stereoisomerism in which the isomers can be interconverted exclusively by rotations about formally single bonds...
change of the enzyme or substrates, such as those involved in the release of product(s) from the enzyme.
Knowledge of the enzyme's structure
Protein structure
Proteins are an important class of biological macromolecules present in all organisms. Proteins are polymers of amino acids. Classified by their physical size, proteins are nanoparticles . Each protein polymer – also known as a polypeptide – consists of a sequence formed from 20 possible L-α-amino...
is helpful in interpreting kinetic data. For example, the structure can suggest how substrates and products bind during catalysis; what changes occur during the reaction; and even the role of particular amino acid
Amino acid
Amino acids are molecules containing an amine group, a carboxylic acid group and a side-chain that varies between different amino acids. The key elements of an amino acid are carbon, hydrogen, oxygen, and nitrogen...
residues in the mechanism. Some enzymes change shape significantly during the mechanism; in such cases, it is helpful to determine the enzyme structure with and without bound substrate analogues that do not undergo the enzymatic reaction.
Not all biological catalysts are protein enzymes; RNA
RNA
Ribonucleic acid , or RNA, is one of the three major macromolecules that are essential for all known forms of life....
-based catalysts such as ribozymes and ribosomes are essential to many cellular functions, such as RNA splicing
Splicing (genetics)
In molecular biology and genetics, splicing is a modification of an RNA after transcription, in which introns are removed and exons are joined. This is needed for the typical eukaryotic messenger RNA before it can be used to produce a correct protein through translation...
and translation. The main difference between ribozymes and enzymes is that RNA catalysts are composed of nucleotides, whereas enzymes are composed of amino acids. Ribozymes also perform a more limited set of reactions, although their reaction mechanism
Reaction mechanism
In chemistry, a reaction mechanism is the step by step sequence of elementary reactions by which overall chemical change occurs.Although only the net chemical change is directly observable for most chemical reactions, experiments can often be designed that suggest the possible sequence of steps in...
s and kinetics can be analysed and classified by the same methods.
General principles
.svg.png)
Chemical equilibrium
In a chemical reaction, chemical equilibrium is the state in which the concentrations of the reactants and products have not yet changed with time. It occurs only in reversible reactions, and not in irreversible reactions. Usually, this state results when the forward reaction proceeds at the same...
between substrates and products. However, unlike uncatalysed chemical reactions, enzyme-catalysed reactions display saturation kinetics. For a given enzyme concentration and for relatively low substrate concentrations, the reaction rate increases linearly with substrate concentration; the enzyme molecules are largely free to catalyse the reaction, and increasing substrate concentration means an increasing rate at which the enzyme and substrate molecules encounter one another. However, at relatively high substrate concentrations, the reaction rate asymptotically
Asymptote
In analytic geometry, an asymptote of a curve is a line such that the distance between the curve and the line approaches zero as they tend to infinity. Some sources include the requirement that the curve may not cross the line infinitely often, but this is unusual for modern authors...
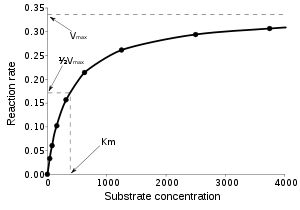
approaches the theoretical maximum; the enzyme active sites are almost all occupied and the reaction rate is determined by the intrinsic turnover rate of the enzyme. The substrate concentration midway between these two limiting cases is denoted by KM.
The two most important kinetic properties of an enzyme are how quickly the enzyme becomes saturated with a particular substrate, and the maximum rate it can achieve. Knowing these properties suggests what an enzyme might do in the cell and can show how the enzyme will respond to changes in these conditions.
Enzyme assays
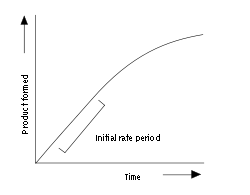
Enzyme assay
Enzyme assays are laboratory methods for measuring enzymatic activity. They are vital for the study of enzyme kinetics and enzyme inhibition.-Enzyme units:...
s are laboratory procedures that measure the rate of enzyme reactions. Because enzymes are not consumed by the reactions they catalyse, enzyme assays usually follow changes in the concentration of either substrates or products to measure the rate of reaction. There are many methods of measurement. Spectrophotometric
Ultraviolet-visible spectroscopy
Ultraviolet-visible spectroscopy or ultraviolet-visible spectrophotometry refers to absorption spectroscopy or reflectance spectroscopy in the ultraviolet-visible spectral region. This means it uses light in the visible and adjacent ranges...
assays observe change in the absorbance of light between products and reactants; radiometric assays involve the incorporation or release of radioactivity to measure the amount of product made over time. Spectrophotometric assays are most convenient since they allow the rate of the reaction to be measured continuously. Although radiometric assays require the removal and counting of samples (i.e., they are discontinuous assays) they are usually extremely sensitive and can measure very low levels of enzyme activity. An analogous approach is to use mass spectrometry
Mass spectrometry
Mass spectrometry is an analytical technique that measures the mass-to-charge ratio of charged particles.It is used for determining masses of particles, for determining the elemental composition of a sample or molecule, and for elucidating the chemical structures of molecules, such as peptides and...
to monitor the incorporation or release of stable isotope
Stable isotope
Stable isotopes are chemical isotopes that may or may not be radioactive, but if radioactive, have half-lives too long to be measured.Only 90 nuclides from the first 40 elements are energetically stable to any kind of decay save proton decay, in theory...
s as substrate is converted into product.
The most sensitive enzyme assays use laser
Laser
A laser is a device that emits light through a process of optical amplification based on the stimulated emission of photons. The term "laser" originated as an acronym for Light Amplification by Stimulated Emission of Radiation...
s focused through a microscope
Microscope
A microscope is an instrument used to see objects that are too small for the naked eye. The science of investigating small objects using such an instrument is called microscopy...
to observe changes in single enzyme molecules as they catalyse their reactions. These measurements either use changes in the fluorescence
Fluorescence
Fluorescence is the emission of light by a substance that has absorbed light or other electromagnetic radiation of a different wavelength. It is a form of luminescence. In most cases, emitted light has a longer wavelength, and therefore lower energy, than the absorbed radiation...
of cofactors
Cofactor (biochemistry)
A cofactor is a non-protein chemical compound that is bound to a protein and is required for the protein's biological activity. These proteins are commonly enzymes, and cofactors can be considered "helper molecules" that assist in biochemical transformations....
during an enzyme's reaction mechanism, or of fluorescent dyes added onto specific sites of the protein
Protein
Proteins are biochemical compounds consisting of one or more polypeptides typically folded into a globular or fibrous form, facilitating a biological function. A polypeptide is a single linear polymer chain of amino acids bonded together by peptide bonds between the carboxyl and amino groups of...
to report movements that occur during catalysis. These studies are providing a new view of the kinetics and dynamics of single enzymes, as opposed to traditional enzyme kinetics, which observes the average behaviour of populations of millions of enzyme molecules.
An example progress curve for an enzyme assay is shown above. The enzyme produces product at an initial rate that is approximately linear for a short period after the start of the reaction. As the reaction proceeds and substrate is consumed, the rate continuously slows (so long as substrate is not still at saturating levels). To measure the initial (and maximal) rate, enzyme assays are typically carried out while the reaction has progressed only a few percent towards total completion. The length of the initial rate period depends on the assay conditions and can range from milliseconds to hours. However, equipment for rapidly mixing liquids allows fast kinetic measurements on initial rates of less than one second. These very rapid assays are essential for measuring pre-steady-state kinetics, which are discussed below.
Most enzyme kinetics studies concentrate on this initial, approximately linear part of enzyme reactions. However, it is also possible to measure the complete reaction curve and fit this data to a non-linear rate equation
Rate equation
The rate law or rate equation for a chemical reaction is an equation that links the reaction rate with concentrations or pressures of reactants and constant parameters . To determine the rate equation for a particular system one combines the reaction rate with a mass balance for the system...
. This way of measuring enzyme reactions is called progress-curve analysis. This approach is useful as an alternative to rapid kinetics when the initial rate is too fast to measure accurately.
Single-substrate reactions
Enzymes with single-substrate mechanisms include isomeraseIsomerase
In biochemistry, an isomerase is an enzyme that catalyzes the structural rearrangement of isomers. Isomerases thus catalyze reactions of the formwhere B is an isomer of A.-Nomenclature:...
s such as triosephosphateisomerase
Triosephosphateisomerase
Triose-phosphate isomerase , is an enzyme that catalyzes the reversible interconversion of the triose phosphate isomers dihydroxyacetone phosphate and D-glyceraldehyde 3-phosphate....
or bisphosphoglycerate mutase
Bisphosphoglycerate mutase
Bisphosphoglycerate mutase is an enzyme unique to erythrocytes and placental cells. It is responsible for the catalytic synthesis of 2,3-Bisphosphoglycerate from 1,3-bisphosphoglycerate...
, intramolecular lyase
Lyase
In biochemistry, a lyase is an enzyme that catalyzes the breaking of various chemical bonds by means other than hydrolysis and oxidation, often forming a new double bond or a new ring structure...
s such as adenylate cyclase
Adenylate cyclase
Adenylate cyclase is part of the G protein signalling cascade, which transmits chemical signals from outside the cell across the membrane to the inside of the cell ....
and the hammerhead ribozyme
Ribozyme
A ribozyme is an RNA molecule with a well defined tertiary structure that enables it to catalyze a chemical reaction. Ribozyme means ribonucleic acid enzyme. It may also be called an RNA enzyme or catalytic RNA. Many natural ribozymes catalyze either the hydrolysis of one of their own...
, a RNA lyase. However, some enzymes that only have a single substrate do not fall into this category of mechanisms. Catalase
Catalase
Catalase is a common enzyme found in nearly all living organisms that are exposed to oxygen, where it catalyzes the decomposition of hydrogen peroxide to water and oxygen...
is an example of this, as the enzyme reacts with a first molecule of hydrogen peroxide
Hydrogen peroxide
Hydrogen peroxide is the simplest peroxide and an oxidizer. Hydrogen peroxide is a clear liquid, slightly more viscous than water. In dilute solution, it appears colorless. With its oxidizing properties, hydrogen peroxide is often used as a bleach or cleaning agent...
substrate, becomes oxidised and is then reduced by a second molecule of substrate. Although a single substrate is involved, the existence of a modified enzyme intermediate means that the mechanism of catalase is actually a ping–pong mechanism, a type of mechanism that is discussed in the Multi-substrate reactions section below.
Michaelis–Menten kinetics

The Michaelis–Menten kinetic model of a single-substrate reaction is shown on the right. There is an initial bimolecular reaction
Chemical kinetics
Chemical kinetics, also known as reaction kinetics, is the study of rates of chemical processes. Chemical kinetics includes investigations of how different experimental conditions can influence the speed of a chemical reaction and yield information about the reaction's mechanism and transition...
between the enzyme E and substrate S to form the enzyme–substrate complex ES. Although the enzymatic mechanism for the unimolecular reaction
Chemical kinetics
Chemical kinetics, also known as reaction kinetics, is the study of rates of chemical processes. Chemical kinetics includes investigations of how different experimental conditions can influence the speed of a chemical reaction and yield information about the reaction's mechanism and transition...
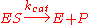 can be quite complex, there is typically one rate-determining enzymatic step that allows this reaction to be modelled as a single catalytic step with an apparent unimolecular rate constant kcat.
can be quite complex, there is typically one rate-determining enzymatic step that allows this reaction to be modelled as a single catalytic step with an apparent unimolecular rate constant kcat.If the reaction path proceeds over one or several intermediates, kcat will be a function of several elementary rate constants, whereas in the simplest case of a single elementary reaction (e.g. no intermediates) it will be identical to the elementary unimolecular rate constant k2. The apparent unimolecular rate constant kcat is also called turnover number and denotes the maximum number of enzymatic reactions catalysed per second.
The Michaelis–Menten equation describes how the (initial) reaction rate v0 depends on the position of the substrate-binding equilibrium
Chemical equilibrium
In a chemical reaction, chemical equilibrium is the state in which the concentrations of the reactants and products have not yet changed with time. It occurs only in reversible reactions, and not in irreversible reactions. Usually, this state results when the forward reaction proceeds at the same...
and the rate constant k2.
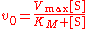 (Michaelis–Menten equation)
(Michaelis–Menten equation)with the constants
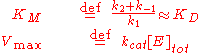
This Michaelis–Menten equation is the basis for most single-substrate enzyme kinetics. Two crucial assumptions underlie this equation (apart from the general assumption about the mechanism only involving no intermediate or product inhibition, and there is no allostericity
Allosteric regulation
In biochemistry, allosteric regulation is the regulation of an enzyme or other protein by binding an effector molecule at the protein's allosteric site . Effectors that enhance the protein's activity are referred to as allosteric activators, whereas those that decrease the protein's activity are...
or cooperativity
Cooperative binding
In biochemistry, a macromolecule exhibits cooperative binding if its affinity for its ligand changes with the amount of ligand already bound.Cooperative binding is a special case of allostery. Cooperative binding requires that the macromolecule have more than one binding site, since cooperativity...
). The first assumption is the so called quasi-steady-state assumption (or pseudo-steady-state hypothesis), namely that the concentration of the substrate-bound enzyme (and hence also the unbound enzyme) changes much more slowly than those of the product and substrate and thus the change over time of the complex can be set to zero
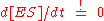 . The second assumption is that the total enzyme concentration does not change over time, thus
. The second assumption is that the total enzyme concentration does not change over time, thus  .
.A complete derivation can be found here.
The Michaelis constant KM is experimentally defined as the concentration at which the rate of the enzyme reaction is half Vmax, which can be verified by substituting [S] = Km into the Michaelis–Menten equation and can also be seen graphically. If the rate-determining enzymatic step is slow compared to substrate dissociation (
 ), the Michaelis constant KM is roughly the dissociation constant
), the Michaelis constant KM is roughly the dissociation constantDissociation constant
In chemistry, biochemistry, and pharmacology, a dissociation constant is a specific type of equilibrium constant that measures the propensity of a larger object to separate reversibly into smaller components, as when a complex falls apart into its component molecules, or when a salt splits up into...
KD of the ES complex.
If
 is small compared to
is small compared to 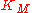 then the term
then the term  and also very little ES complex is formed, thus
and also very little ES complex is formed, thus 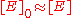 . Therefore, the rate of product formation is
. Therefore, the rate of product formation is
Thus the product formation rate depends on the enzyme concentration as well as on the substrate concentration, the equation resembles a bimolecular reaction with a corresponding pseudo-second order rate constant
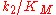 . This constant is a measure of catalytic efficiency. The most efficient enzymes reach a
. This constant is a measure of catalytic efficiency. The most efficient enzymes reach a  in the range of 108 - 1010 M−1 s
in the range of 108 - 1010 M−1 sSecond
The second is a unit of measurement of time, and is the International System of Units base unit of time. It may be measured using a clock....
−1. These enzymes are so efficient they effectively catalyse a reaction each time they encounter a substrate molecule and have thus reached an upper theoretical limit for efficiency (diffusion limit); these enzymes have often been termed perfect enzymes.
Direct use of the Michaelis–Menten equation for time course kinetic analysis
The observed velocities predicted by the Michaelis–Menten equation can be used to directly model the time course disappearance of substrate and the production of product through incorporation of the Michaelis–Menten equation into the equation for first order chemical kinetics. This can only be achieved however if one recognises the problem associated with the use of Euler's number in the description of first order chemical kinetics. i.e. e-k is a split constant that introduces a systematic error into calculations and can be rewritten as a single constant which represents the remaining substrate after each time period.


In 1997, Santiago Schnell
Santiago Schnell
Santiago Schnell is a member of mathematical biology and systems biology community working the field of biochemistry and cellular physiology. He is Associate Professor of , a in the Brehm Center for Type 1 Diabetes Research & Analysis, and a Research Associate Professor of at the University of...
and Claudio Mendoza derived a closed form solution for the time course kinetics analysis of the Michaelis-Menten mechanism. The solution has the form:
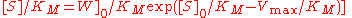
where W[] is the Lambert-W function.
Linear plots of the Michaelis–Menten equation

The plot of v versus [S] above is not linear; although initially linear at low [S], it bends over to saturate at high [S]. Before the modern era of nonlinear curve-fitting
Nonlinear regression
In statistics, nonlinear regression is a form of regression analysis in which observational data are modeled by a function which is a nonlinear combination of the model parameters and depends on one or more independent variables...
on computers, this nonlinearity could make it difficult to estimate KM and Vmax accurately. Therefore, several researchers developed linearisations of the Michaelis–Menten equation, such as the Lineweaver–Burk plot, the Eadie–Hofstee diagram and the Hanes–Woolf plot. All of these linear representations can be useful for visualising data, but none should be used to determine kinetic parameters, as computer software is readily available that allows for more accurate determination by nonlinear regression
Nonlinear regression
In statistics, nonlinear regression is a form of regression analysis in which observational data are modeled by a function which is a nonlinear combination of the model parameters and depends on one or more independent variables...
methods.
The Lineweaver–Burk plot or double reciprocal plot is a common way of illustrating kinetic data. This is produced by taking the reciprocal
Multiplicative inverse
In mathematics, a multiplicative inverse or reciprocal for a number x, denoted by 1/x or x−1, is a number which when multiplied by x yields the multiplicative identity, 1. The multiplicative inverse of a fraction a/b is b/a. For the multiplicative inverse of a real number, divide 1 by the...
of both sides of the Michaelis–Menten equation. As shown on the right, this is a linear form of the Michaelis–Menten equation and produces a straight line with the equation y = mx + c with a y-intercept equivalent to 1/Vmax and an x-intercept of the graph representing -1/KM.

Naturally, no experimental values can be taken at negative 1/[S]; the lower limiting value 1/[S] = 0 (the y-intercept) corresponds to an infinite substrate concentration, where 1/v=1/Vmax as shown at the right; thus, the x-intercept is an extrapolation
Extrapolation
In mathematics, extrapolation is the process of constructing new data points. It is similar to the process of interpolation, which constructs new points between known points, but the results of extrapolations are often less meaningful, and are subject to greater uncertainty. It may also mean...
of the experimental data taken at positive concentrations. More generally, the Lineweaver–Burk plot skews the importance of measurements taken at low substrate concentrations and, thus, can yield inaccurate estimates of Vmax and KM. A more accurate linear plotting method is the Eadie-Hofstee plot
Eadie-Hofstee diagram
In biochemistry, an Eadie–Hofstee diagram is a graphical representation of enzyme kinetics in which reaction velocity is plotted as a function of the velocity vs...
. In this case, v is plotted against v/[S]. In the third common linear representation, the Hanes-Woolf plot
Hanes-Woolf plot
In biochemistry, a Hanes–Woolf plot is a graphical representation of enzyme kinetics in which the ratio of the initial substrate concentration [S] to the reaction velocity v is plotted against [S]...
, [S]/v is plotted against [S].
In general, data normalisation can help diminish the amount of experimental work and can increase the reliability of the output, and is suitable for both graphical and numerical analysis.
Practical significance of kinetic constants
The study of enzyme kinetics is important for two basic reasons. Firstly, it helps explain how enzymes work, and secondly, it helps predict how enzymes behave in living organisms. The kinetic constants defined above, KM and Vmax, are critical to attempts to understand how enzymes work together to control metabolismMetabolism
Metabolism is the set of chemical reactions that happen in the cells of living organisms to sustain life. These processes allow organisms to grow and reproduce, maintain their structures, and respond to their environments. Metabolism is usually divided into two categories...
.
Making these predictions is not trivial, even for simple systems. For example, oxaloacetate is formed by malate dehydrogenase
Malate dehydrogenase
Malate dehydrogenase is an enzyme in the citric acid cycle that catalyzes the conversion of malate into oxaloacetate and vice versa...
within the mitochondrion
Mitochondrion
In cell biology, a mitochondrion is a membrane-enclosed organelle found in most eukaryotic cells. These organelles range from 0.5 to 1.0 micrometers in diameter...
. Oxaloacetate can then be consumed by citrate synthase
Citrate synthase
The enzyme citrate synthase exists in nearly all living cells and stands as a pace-making enzyme in the first step of the Citric Acid Cycle . Citrate synthase is localized within eukaryotic cells in the mitochondrial matrix, but is encoded by nuclear DNA rather than mitochondrial...
, phosphoenolpyruvate carboxykinase
Phosphoenolpyruvate carboxykinase
Phosphoenolpyruvate carboxykinase is an enzyme in the lyase family used in the metabolic pathway of gluconeogenesis. It converts oxaloacetate into phosphoenolpyruvate and carbon dioxide.It is found in two forms, cytosolic and mitochondrial....
or aspartate aminotransferase, feeding into the citric acid cycle
Citric acid cycle
The citric acid cycle — also known as the tricarboxylic acid cycle , the Krebs cycle, or the Szent-Györgyi-Krebs cycle — is a series of chemical reactions which is used by all aerobic living organisms to generate energy through the oxidization of acetate derived from carbohydrates, fats and...
, gluconeogenesis
Gluconeogenesis
Gluconeogenesis is a metabolic pathway that results in the generation of glucose from non-carbohydrate carbon substrates such as lactate, glycerol, and glucogenic amino acids....
or aspartic acid
Aspartic acid
Aspartic acid is an α-amino acid with the chemical formula HOOCCHCH2COOH. The carboxylate anion, salt, or ester of aspartic acid is known as aspartate. The L-isomer of aspartate is one of the 20 proteinogenic amino acids, i.e., the building blocks of proteins...
biosynthesis, respectively. Being able to predict how much oxaloacetate goes into which pathway requires knowledge of the concentration of oxaloacetate as well as the concentration and kinetics of each of these enzymes. This aim of predicting the behaviour of metabolic pathways reaches its most complex expression in the synthesis of huge amounts of kinetic and gene expression
Gene expression
Gene expression is the process by which information from a gene is used in the synthesis of a functional gene product. These products are often proteins, but in non-protein coding genes such as ribosomal RNA , transfer RNA or small nuclear RNA genes, the product is a functional RNA...
data into mathematical models of entire organisms. Alternatively, one useful simplification of the metabolic modelling problem is to ignore the underlying enzyme kinetics and only rely on information about the reaction network's stoichiometry, a technique called flux balance analysis
Flux balance analysis
Flux balance analysis is a mathematical method for analysing metabolism. It does not require knowledge of metabolite concentration or details of the enzyme kinetics of the system...
.
Michaelis–Menten kinetics with intermediate
One could also consider the less simple case-
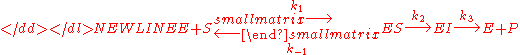
where a complex with the enzyme and an intermediate exists and the intermediate is converted into product in a second step. In this case we have a very similar equation-

but the constants are different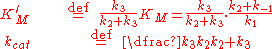
We see that for the limiting case , thus when the last step from EI to E + P is much faster than the previous step, we get again the original equation. Mathematically we have then
, thus when the last step from EI to E + P is much faster than the previous step, we get again the original equation. Mathematically we have then 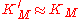 and
and  .
.
Multi-substrate reactions
Multi-substrate reactions follow complex rate equations that describe how the substrates bind and in what sequence. The analysis of these reactions is much simpler if the concentration of substrate A is kept constant and substrate B varied. Under these conditions, the enzyme behaves just like a single-substrate enzyme and a plot of v by [S] gives apparent KM and Vmax constants for substrate B. If a set of these measurements is performed at different fixed concentrations of A, these data can be used to work out what the mechanism of the reaction is. For an enzyme that takes two substrates A and B and turns them into two products P and Q, there are two types of mechanism: ternary complex and ping–pong.
Ternary-complex mechanisms
In these enzymes, both substrates bind to the enzyme at the same time to produce an EAB ternary complex. The order of binding can either be random (in a random mechanism) or substrates have to bind in a particular sequence (in an ordered mechanism). When a set of v by [S] curves (fixed A, varying B) from an enzyme with a ternary-complex mechanism are plotted in a Lineweaver–Burk plot, the set of lines produced will intersect.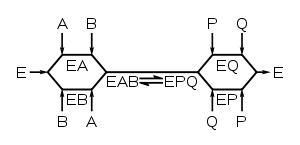
Enzymes with ternary-complex mechanisms include glutathione S-transferaseGlutathione S-transferaseEnzymes of the glutathione S-transferase family are composed of many cytosolic, mitochondrial, and microsomal proteins. GSTs are present in eukaryotes and in prokaryotes, where they catalyze a variety of reactions and accept endogenous and xenobiotic substrates.GSTs can constitute up to 10% of...
, dihydrofolate reductaseDihydrofolate reductase- Function :Dihydrofolate reductase converts dihydrofolate into tetrahydrofolate, a methyl group shuttle required for the de novo synthesis of purines, thymidylic acid, and certain amino acids...
and DNA polymeraseDNA polymeraseA DNA polymerase is an enzyme that helps catalyze in the polymerization of deoxyribonucleotides into a DNA strand. DNA polymerases are best known for their feedback role in DNA replication, in which the polymerase "reads" an intact DNA strand as a template and uses it to synthesize the new strand....
. The following links show short animations of the ternary-complex mechanisms of the enzymes dihydrofolate reductase and DNA polymerase.
Ping–pong mechanisms
As shown on the right, enzymes with a ping-pong mechanism can exist in two states, E and a chemically modified form of the enzyme E*; this modified enzyme is known as an intermediate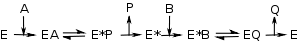 Reactive intermediateIn chemistry a reactive intermediate is a short-lived, high energy, highly reactive molecule. When generated in a chemical reaction it will quickly convert into a more stable molecule. Only in exceptional cases can these compounds be isolated and stored, e.g. low temperatures, matrix isolation...
Reactive intermediateIn chemistry a reactive intermediate is a short-lived, high energy, highly reactive molecule. When generated in a chemical reaction it will quickly convert into a more stable molecule. Only in exceptional cases can these compounds be isolated and stored, e.g. low temperatures, matrix isolation...
. In such mechanisms, substrate A binds, changes the enzyme to E* by, for example, transferring a chemical group to the active site, and is then released. Only after the first substrate is released can substrate B bind and react with the modified enzyme, regenerating the unmodified E form. When a set of v by [S] curves (fixed A, varying B) from an enzyme with a ping–pong mechanism are plotted in a Lineweaver–Burk plot, a set of parallel lines will be produced. This is called a secondary plotSecondary plot (kinetics)In enzyme kinetics, a secondary plot uses the intercept or slope from several Lineweaver-Burk plots to find additional kinetic constants.For example, when a set of v by [S] curves from an enzyme with a ping–pong mechanism are plotted in a Lineweaver–Burk plot, a set of parallel lines will be...
.
Enzymes with ping–pong mechanisms include some oxidoreductases such as thioredoxin peroxidasePeroxidasePeroxidases are a large family of enzymes that typically catalyze a reaction of the form:For many of these enzymes the optimal substrate is hydrogen peroxide, but others are more active with organic hydroperoxides such as lipid peroxides...
, transferases such as acylneuraminate cytydilyltransferase and serine proteaseSerine proteaseSerine proteases are enzymes that cleave peptide bonds in proteins, in which serine serves as the nucleophilic amino acid at the active site.They are found ubiquitously in both eukaryotes and prokaryotes...
s such as trypsinTrypsinTrypsin is a serine protease found in the digestive system of many vertebrates, where it hydrolyses proteins. Trypsin is produced in the pancreas as the inactive proenzyme trypsinogen. Trypsin cleaves peptide chains mainly at the carboxyl side of the amino acids lysine or arginine, except when...
and chymotrypsinChymotrypsinChymotrypsin is a digestive enzyme that can perform proteolysis. Chymotrypsin preferentially cleaves peptide amide bonds where the carboxyl side of the amide bond is a tyrosine, tryptophan, or phenylalanine. These amino acids contain an aromatic ring in their sidechain that fits into a...
. Serine proteases are a very common and diverse family of enzymes, including digestiveDigestionDigestion is the mechanical and chemical breakdown of food into smaller components that are more easily absorbed into a blood stream, for instance. Digestion is a form of catabolism: a breakdown of large food molecules to smaller ones....
enzymes (trypsin, chymotrypsin, and elastase), several enzymes of the blood clotting cascadeCoagulationCoagulation is a complex process by which blood forms clots. It is an important part of hemostasis, the cessation of blood loss from a damaged vessel, wherein a damaged blood vessel wall is covered by a platelet and fibrin-containing clot to stop bleeding and begin repair of the damaged vessel...
and many others. In these serine proteases, the E* intermediate is an acyl-enzyme species formed by the attack of an active site serineSerineSerine is an amino acid with the formula HO2CCHCH2OH. It is one of the proteinogenic amino acids. By virtue of the hydroxyl group, serine is classified as a polar amino acid.-Occurrence and biosynthesis:...
residue on a peptide bondPeptide bondThis article is about the peptide link found within biological molecules, such as proteins. A similar article for synthetic molecules is being created...
in a protein substrate. A short animation showing the mechanism of chymotrypsin is linked here.
Non-Michaelis–Menten kinetics
Some enzymes produce a sigmoid Sigmoid functionMany natural processes, including those of complex system learning curves, exhibit a progression from small beginnings that accelerates and approaches a climax over time. When a detailed description is lacking, a sigmoid function is often used. A sigmoid curve is produced by a mathematical...
Sigmoid functionMany natural processes, including those of complex system learning curves, exhibit a progression from small beginnings that accelerates and approaches a climax over time. When a detailed description is lacking, a sigmoid function is often used. A sigmoid curve is produced by a mathematical...
v by [S] plot, which often indicates cooperative bindingCooperative bindingIn biochemistry, a macromolecule exhibits cooperative binding if its affinity for its ligand changes with the amount of ligand already bound.Cooperative binding is a special case of allostery. Cooperative binding requires that the macromolecule have more than one binding site, since cooperativity...
of substrate to the active site. This means that the binding of one substrate molecule affects the binding of subsequent substrate molecules. This behavior is most common in multimericProtein structureProteins are an important class of biological macromolecules present in all organisms. Proteins are polymers of amino acids. Classified by their physical size, proteins are nanoparticles . Each protein polymer – also known as a polypeptide – consists of a sequence formed from 20 possible L-α-amino...
enzymes with several interacting active sites. Here, the mechanism of cooperation is similar to that of hemoglobinHemoglobinHemoglobin is the iron-containing oxygen-transport metalloprotein in the red blood cells of all vertebrates, with the exception of the fish family Channichthyidae, as well as the tissues of some invertebrates...
, with binding of substrate to one active site altering the affinity of the other active sites for substrate molecules. Positive cooperativity occurs when binding of the first substrate molecule increases the affinity of the other active sites for substrate. Negative cooperativity occurs when binding of the first substrate decreases the affinity of the enzyme for other substrate molecules.
Allosteric enzymes include mammalian tyrosyl tRNA-synthetase, which shows negative cooperativity, and bacterial aspartate transcarbamoylase and phosphofructokinasePhosphofructokinasePhosphofructokinase-1 is the most important regulatory enzyme of glycolysis. It is an allosteric enzyme made of 4 subunits and controlled by many activators and inhibitors...
, which show positive cooperativity.
Cooperativity is surprisingly common and can help regulate the responses of enzymes to changes in the concentrations of their substrates. Positive cooperativity makes enzymes much more sensitive to [S] and their activities can show large changes over a narrow range of substrate concentration. Conversely, negative cooperativity makes enzymes insensitive to small changes in [S].
The Hill equation (biochemistry) is often used to describe the degree of cooperativity quantitatively in non-Michaelis–Menten kinetics. The derived Hill coefficient n measures how much the binding of substrate to one active site affects the binding of substrate to the other active sites. A Hill coefficient of <1 indicates negative cooperativity and a coefficient of >1 indicates positive cooperativityCooperativityCooperativity is a phenomenon displayed by enzymes or receptors that have multiple binding sites where the affinity of the binding sites for a ligand is increased, positive cooperativity, or decreased, negative cooperativity, upon the binding of a ligand to a binding site...
.
Pre-steady-state kinetics
In the first moment after an enzyme is mixed with substrate, no product has been formed and no intermediate Reactive intermediateIn chemistry a reactive intermediate is a short-lived, high energy, highly reactive molecule. When generated in a chemical reaction it will quickly convert into a more stable molecule. Only in exceptional cases can these compounds be isolated and stored, e.g. low temperatures, matrix isolation...
Reactive intermediateIn chemistry a reactive intermediate is a short-lived, high energy, highly reactive molecule. When generated in a chemical reaction it will quickly convert into a more stable molecule. Only in exceptional cases can these compounds be isolated and stored, e.g. low temperatures, matrix isolation...
s exist. The study of the next few milliseconds of the reaction is called Pre-steady-state kinetics also referred to as Burst kineticsBurst kineticsBurst kinetics is a form of enzyme kinetics that refers to an initial high velocity of enzymatic turnover when adding enzyme to substrate. This initial period of high velocity product formation is referred to as the "Burst Phase". This period is observed as the enzymes become saturated with...
. Pre-steady-state kinetics is therefore concerned with the formation and consumption of enzyme–substrate intermediates (such as ES or E*) until their steady-state concentrationsSteady state (chemistry)In chemistry, a steady state is a situation in which all state variables are constant in spite of ongoing processes that strive to change them. For an entire system to be at steady state, i.e. for all state variables of a system to be constant, there must be a flow through the system...
are reached.
This approach was first applied to the hydrolysis reaction catalysed by chymotrypsinChymotrypsinChymotrypsin is a digestive enzyme that can perform proteolysis. Chymotrypsin preferentially cleaves peptide amide bonds where the carboxyl side of the amide bond is a tyrosine, tryptophan, or phenylalanine. These amino acids contain an aromatic ring in their sidechain that fits into a...
. Often, the detection of an intermediate is a vital piece of evidence in investigations of what mechanism an enzyme follows. For example, in the ping–pong mechanisms that are shown above, rapid kinetic measurements can follow the release of product P and measure the formation of the modified enzyme intermediate E*. In the case of chymotrypsin, this intermediate is formed by an attack on the substrate by the nucleophilicNucleophileA nucleophile is a species that donates an electron-pair to an electrophile to form a chemical bond in a reaction. All molecules or ions with a free pair of electrons can act as nucleophiles. Because nucleophiles donate electrons, they are by definition Lewis bases.Nucleophilic describes the...
serine in the active site and the formation of the acyl-enzyme intermediate.
In the figure to the right, the enzyme produces E* rapidly in the first few seconds of the reaction. The rate then slows as steady state is reached. This rapid burst phase of the reaction measures a single turnover of the enzyme. Consequently, the amount of product released in this burst, shown as the intercept on the y-axis of the graph, also gives the amount of functional enzyme which is present in the assay.
Chemical mechanism
An important goal of measuring enzyme kinetics is to determine the chemical mechanism of an enzyme reaction, i.e., the sequence of chemical steps that transform substrate into product. The kinetic approaches discussed above will show at what rates intermediatesReactive intermediateIn chemistry a reactive intermediate is a short-lived, high energy, highly reactive molecule. When generated in a chemical reaction it will quickly convert into a more stable molecule. Only in exceptional cases can these compounds be isolated and stored, e.g. low temperatures, matrix isolation...
are formed and inter-converted, but they cannot identify exactly what these intermediates are.
Kinetic measurements taken under various solution conditions or on slightly modified enzymes or substrates often shed light on this chemical mechanism, as they reveal the rate-determining step or intermediates in the reaction. For example, the breaking of a covalent bondCovalent bondA covalent bond is a form of chemical bonding that is characterized by the sharing of pairs of electrons between atoms. The stable balance of attractive and repulsive forces between atoms when they share electrons is known as covalent bonding....
to a hydrogenHydrogenHydrogen is the chemical element with atomic number 1. It is represented by the symbol H. With an average atomic weight of , hydrogen is the lightest and most abundant chemical element, constituting roughly 75% of the Universe's chemical elemental mass. Stars in the main sequence are mainly...
atomAtomThe atom is a basic unit of matter that consists of a dense central nucleus surrounded by a cloud of negatively charged electrons. The atomic nucleus contains a mix of positively charged protons and electrically neutral neutrons...
is a common rate-determining step. Which of the possible hydrogen transfers is rate determining can be shown by measuring the kinetic effects of substituting each hydrogen by deuteriumDeuteriumDeuterium, also called heavy hydrogen, is one of two stable isotopes of hydrogen. It has a natural abundance in Earth's oceans of about one atom in of hydrogen . Deuterium accounts for approximately 0.0156% of all naturally occurring hydrogen in Earth's oceans, while the most common isotope ...
, its stable isotopeIsotopeIsotopes are variants of atoms of a particular chemical element, which have differing numbers of neutrons. Atoms of a particular element by definition must contain the same number of protons but may have a distinct number of neutrons which differs from atom to atom, without changing the designation...
. The rate will change when the critical hydrogen is replaced, due to a primary kinetic isotope effectKinetic isotope effectThe kinetic isotope effect is the ratio of reaction rates of two different isotopically labeled molecules in a chemical reaction. It is also called "isotope fractionation," although this term is somewhat broader in meaning...
, which occurs because bonds to deuterium are harder to break than bonds to hydrogen. It is also possible to measure similar effects with other isotope substitutions, such as 13C/12C and 18O/16O, but these effects are more subtle.
Isotopes can also be used to reveal the fate of various parts of the substrate molecules in the final products. For example, it is sometimes difficult to discern the origin of an oxygenOxygenOxygen is the element with atomic number 8 and represented by the symbol O. Its name derives from the Greek roots ὀξύς and -γενής , because at the time of naming, it was mistakenly thought that all acids required oxygen in their composition...
atom in the final product; since it may have come from water or from part of the substrate. This may be determined by systematically substituting oxygen's stable isotope 18O into the various molecules that participate in the reaction and checking for the isotope in the product. The chemical mechanism can also be elucidated by examining the kinetics and isotope effects under different pH conditions, by altering the metal ions or other bound cofactorCofactor (biochemistry)A cofactor is a non-protein chemical compound that is bound to a protein and is required for the protein's biological activity. These proteins are commonly enzymes, and cofactors can be considered "helper molecules" that assist in biochemical transformations....
s, by site-directed mutagenesisSite-directed mutagenesisSite-directed mutagenesis, also called site-specific mutagenesis or oligonucleotide-directed mutagenesis, is a molecular biology technique in which a mutation is created at a defined site in a DNA molecule. In general, this form of mutagenesis requires that the wild type gene sequence be known...
of conserved amino acid residues, or by studying the behaviour of the enzyme in the presence of analogues of the substrate(s).
Enzyme inhibition and activation
Enzyme inhibitors are molecules that reduce or abolish enzyme activity, while enzyme activators are molecules that increase the catalytic rate of enzymes. These interactions can be either reversible (i.e., removal of the inhibitor restores enzyme activity) or irreversible (i.e., the inhibitor permanently inactivates the enzyme).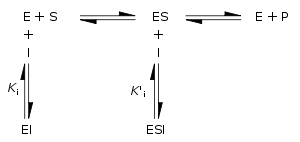
Reversible inhibitors
Traditionally reversible enzyme inhibitors have been classified as competitive, uncompetitive, or non-competitive, according to their effects on Km and Vmax. These different effects result from the inhibitor binding to the enzyme E, to the enzyme–substrate complex ES, or to both, respectively. The particular type of an inhibitor can be discerned by studying the enzyme kinetics as a function of the inhibitor concentration. The three types of inhibition produce Lineweaver–Burke and Eadie–HofsteeEadie-Hofstee diagramIn biochemistry, an Eadie–Hofstee diagram is a graphical representation of enzyme kinetics in which reaction velocity is plotted as a function of the velocity vs...
plots that vary in distinctive ways with inhibitor concentration. For a straightforward qualitative explanation, see
Non-linear regression fits of the enzyme kinetics data to rate equations can yield accurate estimates of dissociation constants and yield important information relating to the mechanism of action.

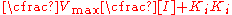
Adding zero to the bottom ([I]-[I])

Dividing by [I]+Ki
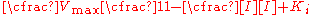

This notation demonstrates that similar to the Michaelis–Menten equation,where the rate of reaction depends on the percent of the enzyme population interacting with substrate
fraction of the enzyme population bound by substrate
fraction of the enzyme population bound by inhibitor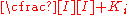
the effect of the inhibitor is a result of the percent of the enzyme population interacting with inhibitor. The only problem with this equation in its present form is that it assumes absolute inhibition of the enzyme with inhibitor binding, when in fact there can be a wide range of affects anywhere from 100% inhibition of substrate turn over to just >0%. To account for this the equation can be easily modified to allow for different degrees of inhibition by including a delta Vmax term.
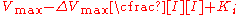
or

This term can then define the residual enzymatic activity present when the inhibitor is interacting with individual enzymes in the population. However the inclusion of this term has the added value of allowing for the possibility of activation if the secondary Vmax term turns out to be higher than the initial term. To account for the possibly of activation as well the notation can then be rewritten replacing the inhibitor "I" with a modifier term denoted here as "X".
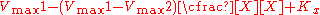
While this terminology results in a simplified way of dealing with kinetic effects relating to the maximum velocity of the Michaelis–Menten equation, it highlights potential problems with the term used to describe effects relating to the Km. The Km relating to the affinity of the enzyme for the substrate should in most cases relate to potential changes in the binding site of the enzyme which would directly result from enzyme inhibitor interactions. As such a term similar to the one proposed above to modulate Vmax should be appropriate in most situations.:
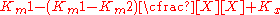
Irreversible inhibitors
Enzyme inhibitors can also irreversibly inactivate enzymes, usually by covalently modifying active site residues. These reactions, which may be called suicide substrates, follow exponential decay functions and are usually saturable. Below saturation, they follow first orderReaction rateThe reaction rate or speed of reaction for a reactant or product in a particular reaction is intuitively defined as how fast or slow a reaction takes place...
kinetics with respect to inhibitor.
Mechanisms of catalysis
The favoured model for the enzyme–substrate interaction is the induced fit model. This model proposes that the initial interaction between enzyme and substrate is relatively weak, but that these weak interactions rapidly induce conformational change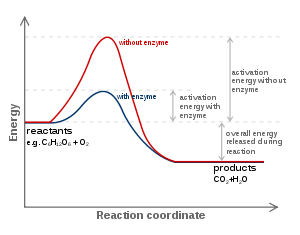 Conformational changeA macromolecule is usually flexible and dynamic. It can change its shape in response to changes in its environment or other factors; each possible shape is called a conformation, and a transition between them is called a conformational change...
Conformational changeA macromolecule is usually flexible and dynamic. It can change its shape in response to changes in its environment or other factors; each possible shape is called a conformation, and a transition between them is called a conformational change...
s in the enzyme that strengthen binding. These conformationalTertiary structureIn biochemistry and molecular biology, the tertiary structure of a protein or any other macromolecule is its three-dimensional structure, as defined by the atomic coordinates.-Relationship to primary structure:...
changes also bring catalytic residues in the active site close to the chemical bonds in the substrate that will be altered in the reaction. Conformational changes can be measured using circular dichroismCircular dichroismCircular dichroism refers to the differential absorption of left and right circularly polarized light. This phenomenon was discovered by Jean-Baptiste Biot, Augustin Fresnel, and Aimé Cotton in the first half of the 19th century. It is exhibited in the absorption bands of optically active chiral...
or dual polarisation interferometryDual Polarisation InterferometryDual polarization interferometry is an analytical technique that can probe molecular scale layers adsorbed to the surface of a waveguide by using the evanescent wave of a laser beam confined to the waveguide...
. After binding takes place, one or more mechanisms of catalysis lower the energy of the reaction's transition stateTransition stateThe transition state of a chemical reaction is a particular configuration along the reaction coordinate. It is defined as the state corresponding to the highest energy along this reaction coordinate. At this point, assuming a perfectly irreversible reaction, colliding reactant molecules will always...
by providing an alternative chemical pathway for the reaction. Mechanisms of catalysis include catalysis by bond strain; by proximity and orientation; by active-site proton donors or acceptors; covalent catalysis and quantum tunnelling.
Enzyme kinetics cannot prove which modes of catalysis are used by an enzyme. However, some kinetic data can suggest possibilities to be examined by other techniques. For example, a ping–pong mechanism with burst-phase pre-steady-state kinetics would suggest covalent catalysis might be important in this enzyme's mechanism. Alternatively, the observation of a strong pH effect on Vmax but not Km might indicate that a residue in the active site needs to be in a particular ionisationIonizationIonization is the process of converting an atom or molecule into an ion by adding or removing charged particles such as electrons or other ions. This is often confused with dissociation. A substance may dissociate without necessarily producing ions. As an example, the molecules of table sugar...
state for catalysis to occur.
Footnotes
α. Link: Interactive Michaelis–Menten kinetics tutorial (Java required)
β. Link: dihydrofolate reductase mechanism (Gif)
γ. Link: DNA polymerase mechanism (Gif)
δ. Link: Chymotrypsin mechanism (Flash required)
External links
- Animation of an enzyme assay — Shows effects of manipulating assay conditions
- MACiE — A database of enzyme reaction mechanisms
- ENZYME — Expasy enzyme nomenclature database
- ENZO — Web application for easy construction and quick testing of kinetic models of enzyme catalyzed reactions.
- ExCatDB — A database of enzyme catalytic mechanisms
- BRENDA — Comprehensive enzyme database, giving substrates, inhibitors and reaction diagrams
- SABIO-RK — A database of reaction kinetics
- Joseph Kraut's Research Group, University of California San Diego — Animations of several enzyme reaction mechanisms
- Symbolism and Terminology in Enzyme Kinetics — A comprehensive explanation of concepts and terminology in enzyme kinetics
- An introduction to enzyme kinetics — An accessible set of on-line tutorials on enzyme kinetics
- Enzyme kinetics animated tutorial — An animated tutorial with audio
-

