LSm
Encyclopedia
RNA
Ribonucleic acid , or RNA, is one of the three major macromolecules that are essential for all known forms of life....
-binding protein
Protein
Proteins are biochemical compounds consisting of one or more polypeptides typically folded into a globular or fibrous form, facilitating a biological function. A polypeptide is a single linear polymer chain of amino acids bonded together by peptide bonds between the carboxyl and amino groups of...
s found in virtually every cellular organism
Organism
In biology, an organism is any contiguous living system . In at least some form, all organisms are capable of response to stimuli, reproduction, growth and development, and maintenance of homoeostasis as a stable whole.An organism may either be unicellular or, as in the case of humans, comprise...
. LSm is a contraction of 'like Sm', because the first identified members of the LSm protein family
Protein family
A protein family is a group of evolutionarily-related proteins, and is often nearly synonymous with gene family. The term protein family should not be confused with family as it is used in taxonomy....
were the Sm proteins. LSm proteins are defined by a characteristic three dimensional structure and their assembly into rings of six or seven individual LSm protein molecule
Molecule
A molecule is an electrically neutral group of at least two atoms held together by covalent chemical bonds. Molecules are distinguished from ions by their electrical charge...
s.
The Sm proteins were first discovered as antigens targeted by so called Anti-Sm antibodies in a patient with a form of systemic lupus erythematosus (SLE)
Lupus erythematosus
Lupus erythematosus is a category for a collection of diseases with similar underlying problems with immunity . Symptoms of these diseases can affect many different body systems, including joints, skin, kidneys, blood cells, heart, and lungs...
, a debilitating autoimmune disease
Autoimmunity
Autoimmunity is the failure of an organism to recognize its own constituent parts as self, which allows an immune response against its own cells and tissues. Any disease that results from such an aberrant immune response is termed an autoimmune disease...
. They were named Sm proteins in honor of this patient, Stephanie Smith. Other proteins with very similar structures were subsequently discovered and named LSm proteins. New members of the LSm protein family continue to be identified and reported.
Proteins with similar structures are grouped into a hierarchy of protein families, superfamilies and folds. The LSm protein structure is an example of a small beta sheet
Beta sheet
The β sheet is the second form of regular secondary structure in proteins, only somewhat less common than the alpha helix. Beta sheets consist of beta strands connected laterally by at least two or three backbone hydrogen bonds, forming a generally twisted, pleated sheet...
folded into a short barrel. Individual LSm proteins assemble into a six or seven member doughnut ring (more properly termed a torus
Torus
In geometry, a torus is a surface of revolution generated by revolving a circle in three dimensional space about an axis coplanar with the circle...
), which usually binds to a small RNA molecule to form a ribonucleoprotein
Ribonucleoprotein
Ribonucleoprotein is a nucleoprotein that contains RNA, i.e. it is an association that combines ribonucleic acid and protein together. A few known examples include the ribosome, the enzyme telomerase, vault ribonucleoproteins, and small nuclear RNPs , which are implicated in pre-mRNA splicing and...
complex. The LSm torus assists the RNA molecule to assume and maintain its proper three dimensional structure. Depending on which LSm proteins and RNA molecule are involved, this ribonucleoprotein complex facilitates a wide variety of RNA processing including degradation, editing, splicing and regulation.
Alternate terms for LSm family are LSm fold and Sm-like fold, and alternate capitalization styles such as lsm and Lsm are common and equally acceptable.
Discovery of the Smith antigen
The story of the discovery of the first LSm proteinProtein
Proteins are biochemical compounds consisting of one or more polypeptides typically folded into a globular or fibrous form, facilitating a biological function. A polypeptide is a single linear polymer chain of amino acids bonded together by peptide bonds between the carboxyl and amino groups of...
s begins with a young woman, Stephanie Smith
Stephanie Smith (artist)
Stephanie Smith , was an aspiring artist from New York City. The anti-Smith antigen found in systemic lupus erythematosus was discovered in her and is the basis of an SLE diagnostic test. She was the sister of New York Times best-selling author, Antonia M. Van-Loon, author of "For Us The...
, who was diagnosed in 1959 with systemic lupus erythematosus (SLE)
Lupus erythematosus
Lupus erythematosus is a category for a collection of diseases with similar underlying problems with immunity . Symptoms of these diseases can affect many different body systems, including joints, skin, kidneys, blood cells, heart, and lungs...
, eventually succumbing to complications of the disease in 1969 at the age of 22. During this period, she was treated at New York's Rockefeller University
Rockefeller University
The Rockefeller University is a private university offering postgraduate and postdoctoral education. It has a strong concentration in the biological sciences. It is also known for producing numerous Nobel laureates...
Hospital, under the care of Dr. Henry Kunkel and Dr. Eng Tan. As an autoimmune disease
Autoimmunity
Autoimmunity is the failure of an organism to recognize its own constituent parts as self, which allows an immune response against its own cells and tissues. Any disease that results from such an aberrant immune response is termed an autoimmune disease...
, SLE patients produce antibodies
Antibody
An antibody, also known as an immunoglobulin, is a large Y-shaped protein used by the immune system to identify and neutralize foreign objects such as bacteria and viruses. The antibody recognizes a unique part of the foreign target, termed an antigen...
to antigen
Antigen
An antigen is a foreign molecule that, when introduced into the body, triggers the production of an antibody by the immune system. The immune system will then kill or neutralize the antigen that is recognized as a foreign and potentially harmful invader. These invaders can be molecules such as...
s in their cells' nuclei
Cell nucleus
In cell biology, the nucleus is a membrane-enclosed organelle found in eukaryotic cells. It contains most of the cell's genetic material, organized as multiple long linear DNA molecules in complex with a large variety of proteins, such as histones, to form chromosomes. The genes within these...
, most frequently to their own DNA
DNA
Deoxyribonucleic acid is a nucleic acid that contains the genetic instructions used in the development and functioning of all known living organisms . The DNA segments that carry this genetic information are called genes, but other DNA sequences have structural purposes, or are involved in...
. However, Dr. Kunkel and Dr. Tan found in 1966 that Ms. Smith produced antibodies
Antibody
An antibody, also known as an immunoglobulin, is a large Y-shaped protein used by the immune system to identify and neutralize foreign objects such as bacteria and viruses. The antibody recognizes a unique part of the foreign target, termed an antigen...
to a set of nuclear proteins, which they named the 'smith antigen
Antigen
An antigen is a foreign molecule that, when introduced into the body, triggers the production of an antibody by the immune system. The immune system will then kill or neutralize the antigen that is recognized as a foreign and potentially harmful invader. These invaders can be molecules such as...
' (Sm Ag). About 30% of SLE patients produce antibodies to these proteins, as opposed to double stranded DNA. This discovery improved diagnostic testing for SLE, but the nature and function of this antigen was unknown.
Sm proteins, snRNPs, the spliceosome and messenger RNA splicing
Research continued during the 1970s and early 1980s. The smith antigen was found to be a complex of ribonucleic acid (RNARNA
Ribonucleic acid , or RNA, is one of the three major macromolecules that are essential for all known forms of life....
) molecules and multiple proteins. A set of uridine
Uridine
Uridine is a molecule that is formed when uracil is attached to a ribose ring via a β-N1-glycosidic bond.If uracil is attached to a deoxyribose ring, it is known as a deoxyuridine....
-rich small nuclear RNA
Non-coding RNA
A non-coding RNA is a functional RNA molecule that is not translated into a protein. Less-frequently used synonyms are non-protein-coding RNA , non-messenger RNA and functional RNA . The term small RNA is often used for short bacterial ncRNAs...
(snRNA) molecules was part of this complex, and given the names U1, U2, U4, U5 and U6. Four of these snRNAs (U1, U2, U4 and U5) were found to be tightly bound to several small proteins, which were named SmB, SmD, SmE, SmF, and SmG in decreasing order of size. SmB has an alternatively spliced variant, SmB', and a very similar protein, SmN replaces SmB'/B in certain (mostly neural) tissues. SmD was later discovered to be a mixture of three proteins, which were named SmD1, SmD2 and SmD3. These nine proteins (SmB, SmB', SmN, SmD1, SmD2, SmD3, SmE, SmF and SmG) became known as the Sm core proteins, or simply Sm proteins. The snRNAs are complexed with the Sm core proteins and with other proteins to form particles in the cell's nucleus
Cell nucleus
In cell biology, the nucleus is a membrane-enclosed organelle found in eukaryotic cells. It contains most of the cell's genetic material, organized as multiple long linear DNA molecules in complex with a large variety of proteins, such as histones, to form chromosomes. The genes within these...
called small nuclear ribonucleoproteins, or snRNP
SnRNP
snRNPs , or small nuclear ribonucleoproteins, are RNA-protein complexes that combine with unmodified pre-mRNA and various other proteins to form a spliceosome, a large RNA-protein molecular complex upon which splicing of pre-mRNA occurs...
s. By the mid 1980s, it became clear that these snRNPs help form a large (4.8 MD molecular weight
Molecular mass
The molecular mass of a substance is the mass of one molecule of that substance, in unified atomic mass unit u...
) complex, called the spliceosome
Spliceosome
A spliceosome is a complex of snRNA and protein subunits that removes introns from a transcribed pre-mRNA segment. This process is generally referred to as splicing.-Composition:...
, around pre-mRNA
Precursor mRNA
Precursor mRNA is an immature single strand of messenger ribonucleic acid . pre-mRNA is synthesized from a DNA template in the cell nucleus by transcription. Pre-mRNA comprises the bulk of heterogeneous nuclear RNA...
, excising portions of the pre-mRNA called intron
Intron
An intron is any nucleotide sequence within a gene that is removed by RNA splicing to generate the final mature RNA product of a gene. The term intron refers to both the DNA sequence within a gene, and the corresponding sequence in RNA transcripts. Sequences that are joined together in the final...
s and splicing the coding portions (exon
Exon
An exon is a nucleic acid sequence that is represented in the mature form of an RNA molecule either after portions of a precursor RNA have been removed by cis-splicing or when two or more precursor RNA molecules have been ligated by trans-splicing. The mature RNA molecule can be a messenger RNA...
s) together. After a few more modifications, the spliced pre-mRNA becomes messenger RNA
Messenger RNA
Messenger RNA is a molecule of RNA encoding a chemical "blueprint" for a protein product. mRNA is transcribed from a DNA template, and carries coding information to the sites of protein synthesis: the ribosomes. Here, the nucleic acid polymer is translated into a polymer of amino acids: a protein...
(mRNA) which is then exported from the nucleus and translated into a protein by ribosomes.
Discovery of proteins similar to the Sm proteins
The snRNA U6 (unlike U1, U2, U4 and U5) does not associate with the Sm proteins, even though the U6 snRNP is a central component in the spliceosomeSpliceosome
A spliceosome is a complex of snRNA and protein subunits that removes introns from a transcribed pre-mRNA segment. This process is generally referred to as splicing.-Composition:...
. In 1999 a protein heteromer was found that binds specifically to U6, and consisted of seven proteins clearly homologous to the Sm proteins. These proteins were denoted LSm (like Sm) proteins (LSm1, LSm2, LSm3, LSm4, LSm5, LSm6 and LSm7), with the similar LSm8 protein identified later. In the bacterium Escherichia coli
Escherichia coli
Escherichia coli is a Gram-negative, rod-shaped bacterium that is commonly found in the lower intestine of warm-blooded organisms . Most E. coli strains are harmless, but some serotypes can cause serious food poisoning in humans, and are occasionally responsible for product recalls...
the Sm-like protein HF-I encoded by the gene hfq was described in 1968 as an essential host factor for RNA bacteriophage
Bacteriophage
A bacteriophage is any one of a number of viruses that infect bacteria. They do this by injecting genetic material, which they carry enclosed in an outer protein capsid...
Qβ replication. The genome
Genome
In modern molecular biology and genetics, the genome is the entirety of an organism's hereditary information. It is encoded either in DNA or, for many types of virus, in RNA. The genome includes both the genes and the non-coding sequences of the DNA/RNA....
of Saccharomyces cerevisiae
Saccharomyces cerevisiae
Saccharomyces cerevisiae is a species of yeast. It is perhaps the most useful yeast, having been instrumental to baking and brewing since ancient times. It is believed that it was originally isolated from the skin of grapes...
(Baker's Yeast) was sequenced in the mid-1990s, providing a rich resource for identifying homologs
Homology (biology)
Homology forms the basis of organization for comparative biology. In 1843, Richard Owen defined homology as "the same organ in different animals under every variety of form and function". Organs as different as a bat's wing, a seal's flipper, a cat's paw and a human hand have a common underlying...
of these human proteins. Subsequently as more eukaryote
Eukaryote
A eukaryote is an organism whose cells contain complex structures enclosed within membranes. Eukaryotes may more formally be referred to as the taxon Eukarya or Eukaryota. The defining membrane-bound structure that sets eukaryotic cells apart from prokaryotic cells is the nucleus, or nuclear...
s genomes were sequenced, it became clear that eukaryotes, in general, share homologs to the same set of seven Sm and eight LSm proteins. Soon after, proteins homologous to these eukaryote LSm proteins were found in Archaea
Archaea
The Archaea are a group of single-celled microorganisms. A single individual or species from this domain is called an archaeon...
(Sm1 and Sm2) and Bacteria
Bacteria
Bacteria are a large domain of prokaryotic microorganisms. Typically a few micrometres in length, bacteria have a wide range of shapes, ranging from spheres to rods and spirals...
(Hfq and YlxS homologs). Interestingly, the archaeal LSm proteins are more similar to the eukaryote LSm proteins than either are to bacterial LSm proteins. The LSm proteins described thus far were rather small proteins, varying from 76 amino acid
Amino acid
Amino acids are molecules containing an amine group, a carboxylic acid group and a side-chain that varies between different amino acids. The key elements of an amino acid are carbon, hydrogen, oxygen, and nitrogen...
s (8.7 kD molecular weight
Molecular mass
The molecular mass of a substance is the mass of one molecule of that substance, in unified atomic mass unit u...
) for human SmG to 231 amino acids (29 kD molecular weight) for human SmB. But recently, larger proteins have been discovered that include a LSm structural domain in addition to other protein structural domains (such as LSm10, LSm11, LSm12, LSm13, LSm14, LSm15, LSm16, ataxin-2, as well as archaeal Sm3).
Discovery of the LSm fold
Around 1995, comparisons between the various LSm homologsHomology (biology)
Homology forms the basis of organization for comparative biology. In 1843, Richard Owen defined homology as "the same organ in different animals under every variety of form and function". Organs as different as a bat's wing, a seal's flipper, a cat's paw and a human hand have a common underlying...
identified two sequence motif
Sequence motif
In genetics, a sequence motif is a nucleotide or amino-acid sequence pattern that is widespread and has, or is conjectured to have, a biological significance...
s, 32 amino acids long and 14 amino acids long, that were very similar in each LSm homolog, and were separated by a non-conserved region of variable length. This indicated the importance of these two sequence motifs (named Sm1 and Sm2), and suggested that all LSm protein genes evolved from a single ancestral gene. In 1999, crystals of recombinant
Recombinant DNA
Recombinant DNA molecules are DNA sequences that result from the use of laboratory methods to bring together genetic material from multiple sources, creating sequences that would not otherwise be found in biological organisms...
Sm proteins were prepared, allowing X-ray crystallography
X-ray crystallography
X-ray crystallography is a method of determining the arrangement of atoms within a crystal, in which a beam of X-rays strikes a crystal and causes the beam of light to spread into many specific directions. From the angles and intensities of these diffracted beams, a crystallographer can produce a...
and determination of their atomic structure in three dimensions. This demonstrated that the LSm proteins share a similar three-dimensional fold of a short alpha helix
Alpha helix
A common motif in the secondary structure of proteins, the alpha helix is a right-handed coiled or spiral conformation, in which every backbone N-H group donates a hydrogen bond to the backbone C=O group of the amino acid four residues earlier...
and a five-stranded folded beta sheet
Beta sheet
The β sheet is the second form of regular secondary structure in proteins, only somewhat less common than the alpha helix. Beta sheets consist of beta strands connected laterally by at least two or three backbone hydrogen bonds, forming a generally twisted, pleated sheet...
, subsequently named the LSm fold. Other investigations found that LSm proteins assemble into a torus
Torus
In geometry, a torus is a surface of revolution generated by revolving a circle in three dimensional space about an axis coplanar with the circle...
(doughnut-shaped ring) of six or seven LSm proteins, and that RNA binds to the inside of the torus, with one nucleotide
Nucleotide
Nucleotides are molecules that, when joined together, make up the structural units of RNA and DNA. In addition, nucleotides participate in cellular signaling , and are incorporated into important cofactors of enzymatic reactions...
bound to each LSm protein.
Characteristics
Uracil
Uracil is one of the four nucleobases in the nucleic acid of RNA that are represented by the letters A, G, C and U. The others are adenine, cytosine, and guanine. In RNA, uracil binds to adenine via two hydrogen bonds. In DNA, the uracil nucleobase is replaced by thymine.Uracil is a common and...
is stacked between the histidine
Histidine
Histidine Histidine, an essential amino acid, has a positively charged imidazole functional group. It is one of the 22 proteinogenic amino acids. Its codons are CAU and CAC. Histidine was first isolated by German physician Albrecht Kossel in 1896. Histidine is an essential amino acid in humans...
and arginine
Arginine
Arginine is an α-amino acid. The L-form is one of the 20 most common natural amino acids. At the level of molecular genetics, in the structure of the messenger ribonucleic acid mRNA, CGU, CGC, CGA, CGG, AGA, and AGG, are the triplets of nucleotide bases or codons that codify for arginine during...
residues, stabilized by hydrogen bond
Hydrogen bond
A hydrogen bond is the attractive interaction of a hydrogen atom with an electronegative atom, such as nitrogen, oxygen or fluorine, that comes from another molecule or chemical group. The hydrogen must be covalently bonded to another electronegative atom to create the bond...
ing to an asparagine
Asparagine
Asparagine is one of the 20 most common natural amino acids on Earth. It has carboxamide as the side-chain's functional group. It is not an essential amino acid...
reside, and hydrogen bond
Hydrogen bond
A hydrogen bond is the attractive interaction of a hydrogen atom with an electronegative atom, such as nitrogen, oxygen or fluorine, that comes from another molecule or chemical group. The hydrogen must be covalently bonded to another electronegative atom to create the bond...
ing between the aspartate
Aspartic acid
Aspartic acid is an α-amino acid with the chemical formula HOOCCHCH2COOH. The carboxylate anion, salt, or ester of aspartic acid is known as aspartate. The L-isomer of aspartate is one of the 20 proteinogenic amino acids, i.e., the building blocks of proteins...
residue and the ribose
Ribose
Ribose is an organic compound with the formula C5H10O5; specifically, a monosaccharide with linear form H––4–H, which has all the hydroxyl groups on the same side in the Fischer projection....
. The lumen of the LSm torus is to the right, and the bulk of the LSm protein is to the left. Only the six amino acid
Amino acid
Amino acids are molecules containing an amine group, a carboxylic acid group and a side-chain that varies between different amino acids. The key elements of an amino acid are carbon, hydrogen, oxygen, and nitrogen...
residues in these two loops are shown for clarity.]]LSm proteins are characterized by a beta sheet
Beta sheet
The β sheet is the second form of regular secondary structure in proteins, only somewhat less common than the alpha helix. Beta sheets consist of beta strands connected laterally by at least two or three backbone hydrogen bonds, forming a generally twisted, pleated sheet...
(the secondary structure
Secondary structure
In biochemistry and structural biology, secondary structure is the general three-dimensional form of local segments of biopolymers such as proteins and nucleic acids...
), folded into the LSm fold (the tertiary structure
Tertiary structure
In biochemistry and molecular biology, the tertiary structure of a protein or any other macromolecule is its three-dimensional structure, as defined by the atomic coordinates.-Relationship to primary structure:...
), polymerization into a six or seven member torus
Torus
In geometry, a torus is a surface of revolution generated by revolving a circle in three dimensional space about an axis coplanar with the circle...
(the quaternary structure
Quaternary structure
In biochemistry, quaternary structure is the arrangement of multiple folded protein or coiling protein molecules in a multi-subunit complex.-Description and examples:...
), and binding to RNA
RNA
Ribonucleic acid , or RNA, is one of the three major macromolecules that are essential for all known forms of life....
oligonucleotides. A modern paradigm classifies proteins on the basis of protein structure
Protein structure
Proteins are an important class of biological macromolecules present in all organisms. Proteins are polymers of amino acids. Classified by their physical size, proteins are nanoparticles . Each protein polymer – also known as a polypeptide – consists of a sequence formed from 20 possible L-α-amino...
and is a currently active field, with three major approaches, SCOP
Structural Classification of Proteins
The Structural Classification of Proteins database is a largely manual classification of protein structural domains based on similarities of their structures and amino acid sequences. A motivation for this classification is to determine the evolutionary relationship between proteins...
(Structural Classification of Proteins), CATH
CATH
The CATH Protein Structure Classification is a semi-automatic, hierarchical classification of protein domains published in 1997 by Christine Orengo, Janet Thornton and their colleagues....
(Class, Architecture, Topology, Homologous superfamily), and FSSP/DALI
Families of structurally similar proteins
Families of Structurally Similar Proteins or FSSP is a database of structurally superimposed proteins generated using the "Distance-matrix ALIgnment" algorithm. The database is helpful for the comparison of protein structures.-External links:*...
(Families of Structually Similar Proteins).
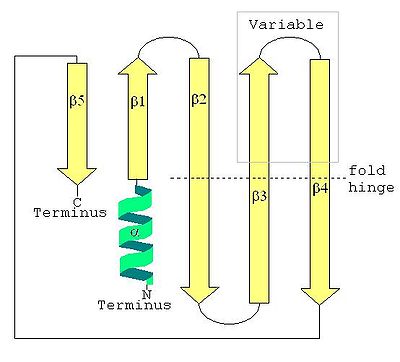
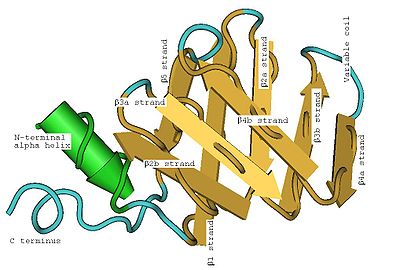
Secondary structure
The secondary structureSecondary structure
In biochemistry and structural biology, secondary structure is the general three-dimensional form of local segments of biopolymers such as proteins and nucleic acids...
of a LSm protein is a small five-strand anti-parallel beta sheet
Beta sheet
The β sheet is the second form of regular secondary structure in proteins, only somewhat less common than the alpha helix. Beta sheets consist of beta strands connected laterally by at least two or three backbone hydrogen bonds, forming a generally twisted, pleated sheet...
, with the strands identified from the N-terminal end
N-terminal end
The N-terminus refers to the start of a protein or polypeptide terminated by an amino acid with a free amine group . The convention for writing peptide sequences is to put the N-terminus on the left and write the sequence from N- to C-terminus...
to the C-terminal end
C-terminal end
The C-terminus is the end of an amino acid chain , terminated by a free carboxyl group . When the protein is translated from messenger RNA, it is created from N-terminus to C-terminus...
as β1, β2, β3, β4, β5. The SCOP class of All beta proteins and the CATH class of Mainly Beta are defined as protein structures that are primarily beta sheets, thus including LSm. The SM1 sequence motif
Sequence motif
In genetics, a sequence motif is a nucleotide or amino-acid sequence pattern that is widespread and has, or is conjectured to have, a biological significance...
corresponds to the β1, β2, β3 strands, and the SM2 sequence motif corresponds to the β4 and β5 strands. The first four beta strands are adjacent to each other, but β5 is adjacent to β1, turning the overall structure into a short barrel. This structural topology is described as 51234. A short (two to four turns) N-terminal alpha helix
Alpha helix
A common motif in the secondary structure of proteins, the alpha helix is a right-handed coiled or spiral conformation, in which every backbone N-H group donates a hydrogen bond to the backbone C=O group of the amino acid four residues earlier...
is also present in most LSm proteins. The β3 and β4 strands are short in some LSm proteins, and are separated by an unstructured coil of variable length. The β2, β3 and β4 strands are strongly bent about 120° degrees at their midpoints The bends in these strands are often glycine
Glycine
Glycine is an organic compound with the formula NH2CH2COOH. Having a hydrogen substituent as its 'side chain', glycine is the smallest of the 20 amino acids commonly found in proteins. Its codons are GGU, GGC, GGA, GGG cf. the genetic code.Glycine is a colourless, sweet-tasting crystalline solid...
, and the side chains internal to the beta barrel are often the hydrophobic residues valine
Valine
Valine is an α-amino acid with the chemical formula HO2CCHCH2. L-Valine is one of 20 proteinogenic amino acids. Its codons are GUU, GUC, GUA, and GUG. This essential amino acid is classified as nonpolar...
, leucine
Leucine
Leucine is a branched-chain α-amino acid with the chemical formula HO2CCHCH2CH2. Leucine is classified as a hydrophobic amino acid due to its aliphatic isobutyl side chain. It is encoded by six codons and is a major component of the subunits in ferritin, astacin and other 'buffer' proteins...
, isoleucine
Isoleucine
Isoleucine is an α-amino acid with the chemical formula HO2CCHCHCH2CH3. It is an essential amino acid, which means that humans cannot synthesize it, so it must be ingested. Its codons are AUU, AUC and AUA....
and methionine
Methionine
Methionine is an α-amino acid with the chemical formula HO2CCHCH2CH2SCH3. This essential amino acid is classified as nonpolar. This amino-acid is coded by the codon AUG, also known as the initiation codon, since it indicates mRNA's coding region where translation into protein...
.
Tertiary structure
SCOP simply classifies the LSm structure as the Sm-like fold, one of 149 different Beta Protein folds, without any intermediate groupings. The LSm beta sheet is sharply bent and described as a Roll architecture in CATH (one of 20 different beta protein architectures in CATH). One of the beta strands (β5 in LSm) crosses the open edge of the roll to form a small SH3 type barrel topology (one of 33 beta roll topologies in CATH). CATH lists 23 homologous superfamilies with an SH3 type barrel topology, one of which is the LSm structure (RNA Binding Protein in the CATH system). SCOP continues its structural classification after Fold to Superfamily, Family and Domain, while CATH continues to Sequence Family, but these divisions are more appropriately described in the "Evolution and phylogeny" section.The SH3-type barrel tertiary structure
Tertiary structure
In biochemistry and molecular biology, the tertiary structure of a protein or any other macromolecule is its three-dimensional structure, as defined by the atomic coordinates.-Relationship to primary structure:...
of the LSm fold is formed by the strongly bent (about 120°) β2, β3 and β4 strands, with the barrel structure closed by the β5 strand. Emphasizing the tertiary structure, each bent beta strand can be described as two shorter beta strands. The LSm fold can be viewed as an eight-strand anti-parallel beta sandwich, with five strands in one plane and three strands in a parallel plane with about a 45° pitch angle between the two halves of the beta sandwich. The short (two to four turns) N-terminal alpha helix
Alpha helix
A common motif in the secondary structure of proteins, the alpha helix is a right-handed coiled or spiral conformation, in which every backbone N-H group donates a hydrogen bond to the backbone C=O group of the amino acid four residues earlier...
occurs at one edge of the beta sandwich. This alpha helix and the beta strands can be labeled (from the N-terminus
N-terminal end
The N-terminus refers to the start of a protein or polypeptide terminated by an amino acid with a free amine group . The convention for writing peptide sequences is to put the N-terminus on the left and write the sequence from N- to C-terminus...
to the C-terminus
C-terminal end
The C-terminus is the end of an amino acid chain , terminated by a free carboxyl group . When the protein is translated from messenger RNA, it is created from N-terminus to C-terminus...
) α, β1, β2a, β2b, β3a, β3b, β4a, β4b, β5 where the a and b refer to either the two halves of a bent strand in the five-strand description, or to the individual strands in the eight-strand description. Each strand (in the eight-strand description) is formed from five amino acid
Amino acid
Amino acids are molecules containing an amine group, a carboxylic acid group and a side-chain that varies between different amino acids. The key elements of an amino acid are carbon, hydrogen, oxygen, and nitrogen...
residues. Including the bends and loops between the strands, and the alpha helix, about 60 amino acid residues contribute to the LSm fold, but this varies between homologs
Homology (biology)
Homology forms the basis of organization for comparative biology. In 1843, Richard Owen defined homology as "the same organ in different animals under every variety of form and function". Organs as different as a bat's wing, a seal's flipper, a cat's paw and a human hand have a common underlying...
due to variation in inter-strand loops, the alpha helix, and even the lengths of β3b and β4a strands.
(Note these images are of the same LSm protein structure in four different views. -->)
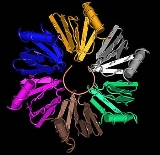
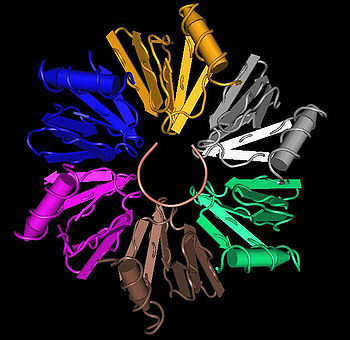
Quaternary structure
LSm proteins typically assemble into a LSm ring, a six or seven member torusTorus
In geometry, a torus is a surface of revolution generated by revolving a circle in three dimensional space about an axis coplanar with the circle...
, about 7 nanometers
Metre
The metre , symbol m, is the base unit of length in the International System of Units . Originally intended to be one ten-millionth of the distance from the Earth's equator to the North Pole , its definition has been periodically refined to reflect growing knowledge of metrology...
in diameter with a 2 nanometer hole. The ancestral condition is a homohexamer
Quaternary structure
In biochemistry, quaternary structure is the arrangement of multiple folded protein or coiling protein molecules in a multi-subunit complex.-Description and examples:...
or homoheptamer
Quaternary structure
In biochemistry, quaternary structure is the arrangement of multiple folded protein or coiling protein molecules in a multi-subunit complex.-Description and examples:...
of identical LSm subunits. LSm proteins in eukaryote
Eukaryote
A eukaryote is an organism whose cells contain complex structures enclosed within membranes. Eukaryotes may more formally be referred to as the taxon Eukarya or Eukaryota. The defining membrane-bound structure that sets eukaryotic cells apart from prokaryotic cells is the nucleus, or nuclear...
s form heteroheptamers
Quaternary structure
In biochemistry, quaternary structure is the arrangement of multiple folded protein or coiling protein molecules in a multi-subunit complex.-Description and examples:...
of seven different LSm subunits, such as the Sm proteins. Binding between the LSm proteins is best understood with the eight-strand description of the LSm fold. The five-strand half of the beta sandwich of one subunit aligns with the three-strand half of the beta sandwich of the adjacent subunit, forming a twisted 8-strand beta sheet Aβ4a/Aβ3b/Aβ2a/Aβ1/Aβ5/Bβ4b/Bβ3a/Bβ2b, where the A and B refer to the two different subunits. In addition to hydrogen bond
Hydrogen bond
A hydrogen bond is the attractive interaction of a hydrogen atom with an electronegative atom, such as nitrogen, oxygen or fluorine, that comes from another molecule or chemical group. The hydrogen must be covalently bonded to another electronegative atom to create the bond...
ing between the Aβ5 and Bβ4b beta strands of the two LSm protein subunits, there are energetically favorable contacts between hydrophobic
Hydrophobic effect
The hydrophobic effect is the observed tendency of nonpolar substances to aggregate in aqueous solution and exclude water molecules. The name, literally meaning "water-fearing," describes the segregation and apparent repulsion between water and nonpolar substances...
amino acid side chains in the interior of the contact area, and energetically favorable contacts between hydrophilic
Hydrophile
A hydrophile, from the Greek "water" and φιλια "love," is a molecule or other molecular entity that is attracted to, and tends to be dissolved by water. A hydrophilic molecule or portion of a molecule is one that has a tendency to interact with or be dissolved by, water and other polar substances...
amino acid side chains around the periphery of the contact area.
RNA oligonucleotide binding
LSm rings form ribonucleoproteinRibonucleoprotein
Ribonucleoprotein is a nucleoprotein that contains RNA, i.e. it is an association that combines ribonucleic acid and protein together. A few known examples include the ribosome, the enzyme telomerase, vault ribonucleoproteins, and small nuclear RNPs , which are implicated in pre-mRNA splicing and...
complexes with RNA
RNA
Ribonucleic acid , or RNA, is one of the three major macromolecules that are essential for all known forms of life....
oligonucleotide
Oligonucleotide
An oligonucleotide is a short nucleic acid polymer, typically with fifty or fewer bases. Although they can be formed by bond cleavage of longer segments, they are now more commonly synthesized, in a sequence-specific manner, from individual nucleoside phosphoramidites...
s that vary in binding strength from very stable complexes (such as the Sm class snRNPs) to transient complexes. Where the details of this binding are known, the RNA oligonucleotides generally bind inside the hole (lumen) of the LSm torus, one nucleotide
Nucleotide
Nucleotides are molecules that, when joined together, make up the structural units of RNA and DNA. In addition, nucleotides participate in cellular signaling , and are incorporated into important cofactors of enzymatic reactions...
per LSm subunit, but additional nucleotide binding sites have been reported at the top (α helix
Alpha helix
A common motif in the secondary structure of proteins, the alpha helix is a right-handed coiled or spiral conformation, in which every backbone N-H group donates a hydrogen bond to the backbone C=O group of the amino acid four residues earlier...
side) of the ring. The exact chemical nature of this binding varies, but common motifs include stacking the heterocyclic base (often uracil
Uracil
Uracil is one of the four nucleobases in the nucleic acid of RNA that are represented by the letters A, G, C and U. The others are adenine, cytosine, and guanine. In RNA, uracil binds to adenine via two hydrogen bonds. In DNA, the uracil nucleobase is replaced by thymine.Uracil is a common and...
) between planar side chains of two amino acids, hydrogen bond
Hydrogen bond
A hydrogen bond is the attractive interaction of a hydrogen atom with an electronegative atom, such as nitrogen, oxygen or fluorine, that comes from another molecule or chemical group. The hydrogen must be covalently bonded to another electronegative atom to create the bond...
ing to the heterocyclic base and/or the ribose
Ribose
Ribose is an organic compound with the formula C5H10O5; specifically, a monosaccharide with linear form H––4–H, which has all the hydroxyl groups on the same side in the Fischer projection....
, and salt bridges
Salt bridge (protein)
Salt bridges fall into the broader category of noncovalent interactions. A salt bridge is actually a combination of two noncovalent interactions: hydrogen bonding and electrostatic interactions . This is most commonly observed to contribute stability to the entropically unfavorable folded...
to the phosphate
Phosphate
A phosphate, an inorganic chemical, is a salt of phosphoric acid. In organic chemistry, a phosphate, or organophosphate, is an ester of phosphoric acid. Organic phosphates are important in biochemistry and biogeochemistry or ecology. Inorganic phosphates are mined to obtain phosphorus for use in...
group.
Functions
The various kinds of LSm rings function as scaffolds or chaperones for RNARNA
Ribonucleic acid , or RNA, is one of the three major macromolecules that are essential for all known forms of life....
oligonucleotide
Oligonucleotide
An oligonucleotide is a short nucleic acid polymer, typically with fifty or fewer bases. Although they can be formed by bond cleavage of longer segments, they are now more commonly synthesized, in a sequence-specific manner, from individual nucleoside phosphoramidites...
s, assisting the RNA to assume and maintain the proper three dimensional structure. In some cases, this allows the oligonucleotide RNA to function catalytically as a ribozyme
Ribozyme
A ribozyme is an RNA molecule with a well defined tertiary structure that enables it to catalyze a chemical reaction. Ribozyme means ribonucleic acid enzyme. It may also be called an RNA enzyme or catalytic RNA. Many natural ribozymes catalyze either the hydrolysis of one of their own...
. In other cases, this facilitates modification or degradation of the RNA, or the assembly, storage, and intracellular transport of ribonucleoprotein
Ribonucleoprotein
Ribonucleoprotein is a nucleoprotein that contains RNA, i.e. it is an association that combines ribonucleic acid and protein together. A few known examples include the ribosome, the enzyme telomerase, vault ribonucleoproteins, and small nuclear RNPs , which are implicated in pre-mRNA splicing and...
complexes.
Sm ring
The Sm ring is found in the nucleusCell nucleus
In cell biology, the nucleus is a membrane-enclosed organelle found in eukaryotic cells. It contains most of the cell's genetic material, organized as multiple long linear DNA molecules in complex with a large variety of proteins, such as histones, to form chromosomes. The genes within these...
of all eukaryote
Eukaryote
A eukaryote is an organism whose cells contain complex structures enclosed within membranes. Eukaryotes may more formally be referred to as the taxon Eukarya or Eukaryota. The defining membrane-bound structure that sets eukaryotic cells apart from prokaryotic cells is the nucleus, or nuclear...
s (about 2.5 x 106 copies per proliferating human cell), and has the best understood functions. The Sm ring is a heteroheptamer
Quaternary structure
In biochemistry, quaternary structure is the arrangement of multiple folded protein or coiling protein molecules in a multi-subunit complex.-Description and examples:...
. The Sm-class snRNA
Non-coding RNA
A non-coding RNA is a functional RNA molecule that is not translated into a protein. Less-frequently used synonyms are non-protein-coding RNA , non-messenger RNA and functional RNA . The term small RNA is often used for short bacterial ncRNAs...
molecule (in the 5' to 3' direction) enters the lumen (doughnut hole) at the SmE subunit and proceeds sequentially in a clockwise fashion (looking from the α helix side) inside the lumen (doughnut hole) to the SmG, SmD3, SmB, SmD1, SmD2 subunits, exiting at the SmF subunit. (SmB can be replaced by the splice variant SmB' and by SmN in neural tissues.) The Sm ring permanently binds to the U1, U2, U4 and U5 snRNAs which form four of the five snRNP
SnRNP
snRNPs , or small nuclear ribonucleoproteins, are RNA-protein complexes that combine with unmodified pre-mRNA and various other proteins to form a spliceosome, a large RNA-protein molecular complex upon which splicing of pre-mRNA occurs...
s that constitute the major spliceosome
Spliceosome
A spliceosome is a complex of snRNA and protein subunits that removes introns from a transcribed pre-mRNA segment. This process is generally referred to as splicing.-Composition:...
. The Sm ring also permanently binds to the U11, U12 and U4atac
U4atac minor spliceosomal RNA
U4atac minor spliceosomal RNA is a ncRNA which is an essential component of the minor U12-type spliceosome complex. The U12-type spliceosome is required for removal of the rarer class of eukaryotic introns ....
snRNAs which form four of the five snRNPs (including the U5 snRNP) that contstitute the minor spliceosome
Minor spliceosome
The minor spliceosome is a ribonucleoprotein complex that catalyses the removal of an atypical class of spliceosomal introns from eukaryotic messenger RNAs in plant, insects, vertebrates and some fungi . This process is called noncanonical splicing, as opposed to U2-dependent canonical splicing...
. Both of these spliceosomes are central RNA-processing complexes in the maturation of messenger RNA
Messenger RNA
Messenger RNA is a molecule of RNA encoding a chemical "blueprint" for a protein product. mRNA is transcribed from a DNA template, and carries coding information to the sites of protein synthesis: the ribosomes. Here, the nucleic acid polymer is translated into a polymer of amino acids: a protein...
from pre-mRNA
Precursor mRNA
Precursor mRNA is an immature single strand of messenger ribonucleic acid . pre-mRNA is synthesized from a DNA template in the cell nucleus by transcription. Pre-mRNA comprises the bulk of heterogeneous nuclear RNA...
. Sm proteins have also been reported to be part of ribonucleoprotein
Ribonucleoprotein
Ribonucleoprotein is a nucleoprotein that contains RNA, i.e. it is an association that combines ribonucleic acid and protein together. A few known examples include the ribosome, the enzyme telomerase, vault ribonucleoproteins, and small nuclear RNPs , which are implicated in pre-mRNA splicing and...
component of telomerase
Telomerase
Telomerase is an enzyme that adds DNA sequence repeats to the 3' end of DNA strands in the telomere regions, which are found at the ends of eukaryotic chromosomes. This region of repeated nucleotide called telomeres contains non-coding DNA material and prevents constant loss of important DNA from...
.
Lsm2-8 ring
The two Lsm2-8 snRNPs (U6 and U6atacU6atac minor spliceosomal RNA
U6atac minor spliceosomal RNA is a non-coding RNA which is an essential component of the minor U12-type spliceosome complex. The U12-type spliceosome is required for removal of the rarer class of eukaryotic introns ....
) have the key catalyic function in the major and minor spliceosomes. These snRNPs do not include the Sm ring, but instead use the heteroheptameric
Quaternary structure
In biochemistry, quaternary structure is the arrangement of multiple folded protein or coiling protein molecules in a multi-subunit complex.-Description and examples:...
Lsm2-8 ring. The LSm rings are about 20 times less abundant than the Sm rings. The order of these seven LSm proteins in this ring is not known, but based on amino acid sequence homology
Homology (biology)
Homology forms the basis of organization for comparative biology. In 1843, Richard Owen defined homology as "the same organ in different animals under every variety of form and function". Organs as different as a bat's wing, a seal's flipper, a cat's paw and a human hand have a common underlying...
with the Sm proteins, it is speculated that the snRNA (in the 5' to 3' direction) may bind first to LSm5, and precedes sequentially clockwise to the LSm7, LSm4, LSm8, LSm2, LSm3, and exiting at the LSm6 subunit. Experiments with Saccharomyces cerevisiae
Saccharomyces cerevisiae
Saccharomyces cerevisiae is a species of yeast. It is perhaps the most useful yeast, having been instrumental to baking and brewing since ancient times. It is believed that it was originally isolated from the skin of grapes...
(budding yeast) mutations suggest that the Lsm2-8 ring assists the reassociation of the U4 and U6 snRNPs into the U4/U6 di-snRNP. (After completion of exon deletion and intron splicing, these two snRNPs must reassociate for the spliceosome to initiate another exon/intron splicing cycle. In this role, the Lsm2-8 ring acts as an RNA chaperone instead of an RNA scaffold.) The Lsm2-8 ring also forms an snRNP with the U8 small nucleolar RNA
Non-coding RNA
A non-coding RNA is a functional RNA molecule that is not translated into a protein. Less-frequently used synonyms are non-protein-coding RNA , non-messenger RNA and functional RNA . The term small RNA is often used for short bacterial ncRNAs...
(snoRNA) which localizes in the nucleolus
Nucleolus
The nucleolus is a non-membrane bound structure composed of proteins and nucleic acids found within the nucleus. Ribosomal RNA is transcribed and assembled within the nucleolus...
. This ribonucleoprotein complex is necessary for processing ribosomal RNA
Ribosomal RNA
Ribosomal ribonucleic acid is the RNA component of the ribosome, the enzyme that is the site of protein synthesis in all living cells. Ribosomal RNA provides a mechanism for decoding mRNA into amino acids and interacts with tRNAs during translation by providing peptidyl transferase activity...
and transfer RNA
Transfer RNA
Transfer RNA is an adaptor molecule composed of RNA, typically 73 to 93 nucleotides in length, that is used in biology to bridge the three-letter genetic code in messenger RNA with the twenty-letter code of amino acids in proteins. The role of tRNA as an adaptor is best understood by...
to their mature forms. The Lsm2-8 ring is reported to have a role in the processing of pre-P RNA into RNase P RNA
RNase P
Ribonuclease P is a type of ribonuclease which cleaves RNA. RNase P is unique from other RNases in that it is a ribozyme – a ribonucleic acid that acts as a catalyst in the same way that a protein based enzyme would. Its function is to cleave off an extra, or precursor, sequence of RNA on tRNA...
. In contrast to the Sm ring, the Lsm2-8 ring does not permanently bind to its snRNA and snoRNA.
Sm10/Sm11 ring
A second type of Sm ring exists where LSm10 replace SmD1 and LSm11 replaces SmD2. LSm11 is a two domain protein with the C-terminalC-terminal end
The C-terminus is the end of an amino acid chain , terminated by a free carboxyl group . When the protein is translated from messenger RNA, it is created from N-terminus to C-terminus...
domain being a LSm domain. This heteroheptamer ring binds with the U7 snRNA in the U7 snRNP. The U7 snRNP mediates processing of the 3'-end of the histone
Histone
In biology, histones are highly alkaline proteins found in eukaryotic cell nuclei that package and order the DNA into structural units called nucleosomes. They are the chief protein components of chromatin, acting as spools around which DNA winds, and play a role in gene regulation...
mRNA
Messenger RNA
Messenger RNA is a molecule of RNA encoding a chemical "blueprint" for a protein product. mRNA is transcribed from a DNA template, and carries coding information to the sites of protein synthesis: the ribosomes. Here, the nucleic acid polymer is translated into a polymer of amino acids: a protein...
in the nucleus. Like the Sm ring, it is assembled in the cytoplasm onto the U7 snRNA by a specialized SMN complex.
Lsm1-7 ring
A second type of Lsm ring is the Lsm1-7 ring, which has the same structure as the Lsm2-8 ring except that LSm1 replaces LSm8. In contrast to the Lsm2-8 ring, the Lsm1-7 ring localizes in the cytoplasmCytoplasm
The cytoplasm is a small gel-like substance residing between the cell membrane holding all the cell's internal sub-structures , except for the nucleus. All the contents of the cells of prokaryote organisms are contained within the cytoplasm...
where it assists in degrading messenger RNA
Messenger RNA
Messenger RNA is a molecule of RNA encoding a chemical "blueprint" for a protein product. mRNA is transcribed from a DNA template, and carries coding information to the sites of protein synthesis: the ribosomes. Here, the nucleic acid polymer is translated into a polymer of amino acids: a protein...
in ribonucleoprotein
Ribonucleoprotein
Ribonucleoprotein is a nucleoprotein that contains RNA, i.e. it is an association that combines ribonucleic acid and protein together. A few known examples include the ribosome, the enzyme telomerase, vault ribonucleoproteins, and small nuclear RNPs , which are implicated in pre-mRNA splicing and...
complexes. This process controls the turnover of messenger RNA so that ribosomal translation of mRNA to protein responds quickly to changes in transcription
Transcription (genetics)
Transcription is the process of creating a complementary RNA copy of a sequence of DNA. Both RNA and DNA are nucleic acids, which use base pairs of nucleotides as a complementary language that can be converted back and forth from DNA to RNA by the action of the correct enzymes...
of DNA to messenger RNA by the cell.
Gemin6 and Gemin7
The SMN complex (described under "Biogenesis of snRNPs") is composed of the SMN protein and other proteins, Gemin2-8. Two of these, Gemin 6 and Gemin7 have been discovered to have the LSm structure, and to form a heterodimer. These may have a chaperone function in the SMN complex to assist the formation of the Sm ring on the Sm-class snRNAsNon-coding RNA
A non-coding RNA is a functional RNA molecule that is not translated into a protein. Less-frequently used synonyms are non-protein-coding RNA , non-messenger RNA and functional RNA . The term small RNA is often used for short bacterial ncRNAs...
.
LSm12-16 and other multi-domain LSm proteins
The LSm12-16 proteins have been described very recently. These are two-domain proteins with a N-terminalN-terminal end
The N-terminus refers to the start of a protein or polypeptide terminated by an amino acid with a free amine group . The convention for writing peptide sequences is to put the N-terminus on the left and write the sequence from N- to C-terminus...
LSm domain and a C-terminal
C-terminal end
The C-terminus is the end of an amino acid chain , terminated by a free carboxyl group . When the protein is translated from messenger RNA, it is created from N-terminus to C-terminus...
methyl transferase domain. Very little is known about the function of these proteins, but presumably they are member of LSm-domain rings that interact with RNA. There is some evidence that LSm12 is possibly involved in mRNA degradation and LSm13-16 may have roles in regulation of mitosis
Mitosis
Mitosis is the process by which a eukaryotic cell separates the chromosomes in its cell nucleus into two identical sets, in two separate nuclei. It is generally followed immediately by cytokinesis, which divides the nuclei, cytoplasm, organelles and cell membrane into two cells containing roughly...
. A large protein of unknown function, ataxin-2, associated with the neurodegenerative disease spinocerebellar ataxia type 2
Spinocerebellar ataxia
Spinocerebellar ataxia is a progressive, degenerative, genetic disease with multiple types, each of which could be considered a disease in its own right.-Classification:...
, also has a N-terminal LSm domain.
Archaeal Sm rings
Two LSm proteins are found in a second domainDomain (biology)
In biological taxonomy, a domain is the highest taxonomic rank of organisms, higher than a kingdom. According to the three-domain system of Carl Woese, introduced in 1990, the Tree of Life consists of three domains: Archaea, Bacteria and Eukarya...
of life, the Archaea
Archaea
The Archaea are a group of single-celled microorganisms. A single individual or species from this domain is called an archaeon...
. These are the Sm1 and Sm2 proteins (not to be confused with the Sm1 and Sm2 sequence motif
Sequence motif
In genetics, a sequence motif is a nucleotide or amino-acid sequence pattern that is widespread and has, or is conjectured to have, a biological significance...
s), and are sometimes identified as Sm-like archaeal proteins SmAP1 and SmAP2 for this reason. Sm1 and Sm2 generally form homoheptamer
Quaternary structure
In biochemistry, quaternary structure is the arrangement of multiple folded protein or coiling protein molecules in a multi-subunit complex.-Description and examples:...
rings, although homohexamer rings have been observed. Sm1 rings are similar to eukaryote
Eukaryote
A eukaryote is an organism whose cells contain complex structures enclosed within membranes. Eukaryotes may more formally be referred to as the taxon Eukarya or Eukaryota. The defining membrane-bound structure that sets eukaryotic cells apart from prokaryotic cells is the nucleus, or nuclear...
Lsm rings in that they form in the absence of RNA while Sm2 rings are similar to eukaryote
Eukaryote
A eukaryote is an organism whose cells contain complex structures enclosed within membranes. Eukaryotes may more formally be referred to as the taxon Eukarya or Eukaryota. The defining membrane-bound structure that sets eukaryotic cells apart from prokaryotic cells is the nucleus, or nuclear...
Sm rings in that they require uridine
Uridine
Uridine is a molecule that is formed when uracil is attached to a ribose ring via a β-N1-glycosidic bond.If uracil is attached to a deoxyribose ring, it is known as a deoxyuridine....
-rich RNA for their formation. They have been reported to associate with RNase P RNA
RNase P
Ribonuclease P is a type of ribonuclease which cleaves RNA. RNase P is unique from other RNases in that it is a ribozyme – a ribonucleic acid that acts as a catalyst in the same way that a protein based enzyme would. Its function is to cleave off an extra, or precursor, sequence of RNA on tRNA...
, suggesting a role in transfer RNA
Transfer RNA
Transfer RNA is an adaptor molecule composed of RNA, typically 73 to 93 nucleotides in length, that is used in biology to bridge the three-letter genetic code in messenger RNA with the twenty-letter code of amino acids in proteins. The role of tRNA as an adaptor is best understood by...
processing, but their function in archaea in this process (and possibly processing other RNA such as ribosomal RNA
Ribosomal RNA
Ribosomal ribonucleic acid is the RNA component of the ribosome, the enzyme that is the site of protein synthesis in all living cells. Ribosomal RNA provides a mechanism for decoding mRNA into amino acids and interacts with tRNAs during translation by providing peptidyl transferase activity...
) is mostly unknown. One of the two main branches of archaea, the crenarchaeotes
Crenarchaeota
In taxonomy, the Crenarchaeota has been classified as either a phylum of the Archaea kingdom or a kingdom of its own...
have a third known type of archaeal LSm protein, Sm3. This is a two-domain protein with a N-terminal
N-terminal end
The N-terminus refers to the start of a protein or polypeptide terminated by an amino acid with a free amine group . The convention for writing peptide sequences is to put the N-terminus on the left and write the sequence from N- to C-terminus...
LSm domain that forms a homoheptamer
Quaternary structure
In biochemistry, quaternary structure is the arrangement of multiple folded protein or coiling protein molecules in a multi-subunit complex.-Description and examples:...
ring. Nothing is known about the function of this LSm protein, but presumably it interacts with, and probably help process, RNA in these organisms.
Bacterial LSm rings
Several LSm proteins have been reported in the third domainDomain (biology)
In biological taxonomy, a domain is the highest taxonomic rank of organisms, higher than a kingdom. According to the three-domain system of Carl Woese, introduced in 1990, the Tree of Life consists of three domains: Archaea, Bacteria and Eukarya...
of life, the Bacteria
Bacteria
Bacteria are a large domain of prokaryotic microorganisms. Typically a few micrometres in length, bacteria have a wide range of shapes, ranging from spheres to rods and spirals...
. Hfq protein forms homohexamer
Quaternary structure
In biochemistry, quaternary structure is the arrangement of multiple folded protein or coiling protein molecules in a multi-subunit complex.-Description and examples:...
rings, and was originally discovered as necessary for infection by the bacteriophage Qβ
Bacteriophage Qβ
Bacteriophage Qβ is an icosahedral virus with a diameter of 25 nm. Its host is Escherichia coli. Qβ enters its host cell through the side of the F pilus.-Genetics:...
, although this is clearly not the native function of this protein in bacteria. It is not universally present in all bacteria, but has been found in Proteobacteria
Proteobacteria
The Proteobacteria are a major group of bacteria. They include a wide variety of pathogens, such as Escherichia, Salmonella, Vibrio, Helicobacter, and many other notable genera....
, Firmicutes
Firmicutes
The Firmicutes are a phylum of bacteria, most of which have Gram-positive cell wall structure. A few, however, such as Megasphaera, Pectinatus, Selenomonas and Zymophilus, have a porous pseudo-outer-membrane that causes them to stain Gram-negative...
, Spirochaetes, Thermotogae
Thermotogae
Thermotogae is a phylum of the domain "Bacteria". This phylum comprises merely the class "Thermotogae", with the order "Thermotogales" and the family "Thermotogaceae"....
, Aquificae
Aquificae
The Aquificae phylum is a diverse collection of bacteria that live in harsh environmental settings. They have been found in hot springs, sulfur pools, and thermal ocean vents. Members of the genus Aquifex, for example, are productive in water between 85 to 95 °C. They are the dominant members of...
and one species of Archaea
Archaea
The Archaea are a group of single-celled microorganisms. A single individual or species from this domain is called an archaeon...
. (This last instance is probably a case of horizontal gene transfer
Horizontal gene transfer
Horizontal gene transfer , also lateral gene transfer , is any process in which an organism incorporates genetic material from another organism without being the offspring of that organism...
.) Hfq is pleiotropic
Pleiotropy
Pleiotropy occurs when one gene influences multiple phenotypic traits. Consequently, a mutation in a pleiotropic gene may have an effect on some or all traits simultaneously...
with a variety of interactions, generally associated with translation regulation. These include blocking ribosome binding to mRNA
Messenger RNA
Messenger RNA is a molecule of RNA encoding a chemical "blueprint" for a protein product. mRNA is transcribed from a DNA template, and carries coding information to the sites of protein synthesis: the ribosomes. Here, the nucleic acid polymer is translated into a polymer of amino acids: a protein...
, marking mRNA for degradation by binding to their poly-A tails
Polyadenylation
Polyadenylation is the addition of a poly tail to an RNA molecule. The poly tail consists of multiple adenosine monophosphates; in other words, it is a stretch of RNA that has only adenine bases. In eukaryotes, polyadenylation is part of the process that produces mature messenger RNA for translation...
, and association with bacterial small regulatory RNAs (such as DsrA RNA) that control translation by binding to certain mRNAs
Messenger RNA
Messenger RNA is a molecule of RNA encoding a chemical "blueprint" for a protein product. mRNA is transcribed from a DNA template, and carries coding information to the sites of protein synthesis: the ribosomes. Here, the nucleic acid polymer is translated into a polymer of amino acids: a protein...
. A second bacterial LSm protein is YlxS (sometimes also called YhbC), which was first identified in the soil bacterium Bacillus subtilis
Bacillus subtilis
Bacillus subtilis, known also as the hay bacillus or grass bacillus, is a Gram-positive, catalase-positive bacterium commonly found in soil. A member of the genus Bacillus, B. subtilis is rod-shaped, and has the ability to form a tough, protective endospore, allowing the organism to tolerate...
. This is a two-domain protein with a N-terminal
N-terminal end
The N-terminus refers to the start of a protein or polypeptide terminated by an amino acid with a free amine group . The convention for writing peptide sequences is to put the N-terminus on the left and write the sequence from N- to C-terminus...
LSm domain. Its function is unknown, but amino acid sequence homologs
Homology (biology)
Homology forms the basis of organization for comparative biology. In 1843, Richard Owen defined homology as "the same organ in different animals under every variety of form and function". Organs as different as a bat's wing, a seal's flipper, a cat's paw and a human hand have a common underlying...
are found in virtually every bacterial genome
Genome
In modern molecular biology and genetics, the genome is the entirety of an organism's hereditary information. It is encoded either in DNA or, for many types of virus, in RNA. The genome includes both the genes and the non-coding sequences of the DNA/RNA....
to date, and it may be an essential protein. The middle domain of the small conductance mechanosensitive channel MscS in Escherichia coli
Escherichia coli
Escherichia coli is a Gram-negative, rod-shaped bacterium that is commonly found in the lower intestine of warm-blooded organisms . Most E. coli strains are harmless, but some serotypes can cause serious food poisoning in humans, and are occasionally responsible for product recalls...
forms a homoheptameric ring. This LSm domain has no apparent RNA-binding function, but the homoheptameric torus is part of the central channel of this membrane protein.
Evolution and phylogeny
LSm homologsHomology (biology)
Homology forms the basis of organization for comparative biology. In 1843, Richard Owen defined homology as "the same organ in different animals under every variety of form and function". Organs as different as a bat's wing, a seal's flipper, a cat's paw and a human hand have a common underlying...
are found in all three domains
Domain (biology)
In biological taxonomy, a domain is the highest taxonomic rank of organisms, higher than a kingdom. According to the three-domain system of Carl Woese, introduced in 1990, the Tree of Life consists of three domains: Archaea, Bacteria and Eukarya...
of life, and may even be found in every single organism
Organism
In biology, an organism is any contiguous living system . In at least some form, all organisms are capable of response to stimuli, reproduction, growth and development, and maintenance of homoeostasis as a stable whole.An organism may either be unicellular or, as in the case of humans, comprise...
. Computational phylogenetic
Computational phylogenetics
Computational phylogenetics is the application of computational algorithms, methods and programs to phylogenetic analyses. The goal is to assemble a phylogenetic tree representing a hypothesis about the evolutionary ancestry of a set of genes, species, or other taxa...
methods are used to infer phylogenetic
Phylogenetics
In biology, phylogenetics is the study of evolutionary relatedness among groups of organisms , which is discovered through molecular sequencing data and morphological data matrices...
relations. Sequence alignment
Sequence alignment
In bioinformatics, a sequence alignment is a way of arranging the sequences of DNA, RNA, or protein to identify regions of similarity that may be a consequence of functional, structural, or evolutionary relationships between the sequences. Aligned sequences of nucleotide or amino acid residues are...
between the various LSm homologs are the appropriate tool for this, such as multiple sequence alignment
Multiple sequence alignment
A multiple sequence alignment is a sequence alignment of three or more biological sequences, generally protein, DNA, or RNA. In many cases, the input set of query sequences are assumed to have an evolutionary relationship by which they share a lineage and are descended from a common ancestor...
of the primary structure (amino acid sequence), and structural alignment
Structural alignment
Structural alignment attempts to establish homology between two or more polymer structures based on their shape and three-dimensional conformation. This process is usually applied to protein tertiary structures but can also be used for large RNA molecules...
of the tertiary structure (three dimensional structure). It is hypothesized that a gene for a LSm protein was present in the last universal ancestor
Last universal ancestor
The last universal ancestor , also called the last universal common ancestor , or the cenancestor, is the most recent organism from which all organisms now living on Earth descend. Thus it is the most recent common ancestor of all current life on Earth...
of all life. Based on the functions of known LSm proteins, this original LSm protein may have assisted ribozyme
Ribozyme
A ribozyme is an RNA molecule with a well defined tertiary structure that enables it to catalyze a chemical reaction. Ribozyme means ribonucleic acid enzyme. It may also be called an RNA enzyme or catalytic RNA. Many natural ribozymes catalyze either the hydrolysis of one of their own...
s in the processing of RNA for synthesizing proteins as part of the RNA world hypothesis
RNA world hypothesis
The RNA world hypothesis proposes that life based on ribonucleic acid pre-dates the current world of life based on deoxyribonucleic acid , RNA and proteins. RNA is able both to store genetic information, like DNA, and to catalyze chemical reactions, like an enzyme protein...
of early life. According to this view, this gene was passed from ancestor to descendent, with frequent mutation
Mutation
In molecular biology and genetics, mutations are changes in a genomic sequence: the DNA sequence of a cell's genome or the DNA or RNA sequence of a virus. They can be defined as sudden and spontaneous changes in the cell. Mutations are caused by radiation, viruses, transposons and mutagenic...
s, gene duplication
Gene duplication
Gene duplication is any duplication of a region of DNA that contains a gene; it may occur as an error in homologous recombination, a retrotransposition event, or duplication of an entire chromosome.The second copy of the gene is often free from selective pressure — that is, mutations of it have no...
s and occasional horizontal gene transfer
Horizontal gene transfer
Horizontal gene transfer , also lateral gene transfer , is any process in which an organism incorporates genetic material from another organism without being the offspring of that organism...
s. In principle, this process can be summarized in a phylogenetic tree
Phylogenetic tree
A phylogenetic tree or evolutionary tree is a branching diagram or "tree" showing the inferred evolutionary relationships among various biological species or other entities based upon similarities and differences in their physical and/or genetic characteristics...
with the root in the last universal ancestor (or earlier), and with the tips representing the universe of LSm genes existing today.
Homomeric LSm rings in bacteria and archaea
Based on structure, the known LSm proteins divide into a group consisting of the bacterial LSm proteins (Hfq, YlxS and MscS) and a second group of all other LSm proteins, in accordance with the most recently published phylogenetic treePhylogenetic tree
A phylogenetic tree or evolutionary tree is a branching diagram or "tree" showing the inferred evolutionary relationships among various biological species or other entities based upon similarities and differences in their physical and/or genetic characteristics...
s. The three archaeal LSm proteins (Sm1, Sm2 and Sm3) also cluster as a group, distinct from the eukaryote LSm proteins. Both the bacterial and archaeal LSm proteins polymerize to homomeric rings, which is the ancestral condition.
Heteromeric LSm rings in eukaryotes
A series of gene duplications of a single eukaryote LSm gene resulted in most (if not all) of the known eukaryote LSm genes. Each of the seven Sm proteins has greater amino acid sequence homologyHomology (biology)
Homology forms the basis of organization for comparative biology. In 1843, Richard Owen defined homology as "the same organ in different animals under every variety of form and function". Organs as different as a bat's wing, a seal's flipper, a cat's paw and a human hand have a common underlying...
to a corresponding Lsm protein than to the other Sm proteins. This suggests that an ancestral LSm gene duplicated several times, resulting in seven paralogs
Homology (biology)
Homology forms the basis of organization for comparative biology. In 1843, Richard Owen defined homology as "the same organ in different animals under every variety of form and function". Organs as different as a bat's wing, a seal's flipper, a cat's paw and a human hand have a common underlying...
. These subsequently diverged from each other so that the ancestral homoheptamer LSm ring became a heteroheptamer ring. Based on the known functions of LSm proteins in eukaryotes and archaea, the ancestral function may have been processing of pre-ribosomal RNA
Ribosomal RNA
Ribosomal ribonucleic acid is the RNA component of the ribosome, the enzyme that is the site of protein synthesis in all living cells. Ribosomal RNA provides a mechanism for decoding mRNA into amino acids and interacts with tRNAs during translation by providing peptidyl transferase activity...
, pre-transfer RNA
Transfer RNA
Transfer RNA is an adaptor molecule composed of RNA, typically 73 to 93 nucleotides in length, that is used in biology to bridge the three-letter genetic code in messenger RNA with the twenty-letter code of amino acids in proteins. The role of tRNA as an adaptor is best understood by...
, and pre-RNase P
RNase P
Ribonuclease P is a type of ribonuclease which cleaves RNA. RNase P is unique from other RNases in that it is a ribozyme – a ribonucleic acid that acts as a catalyst in the same way that a protein based enzyme would. Its function is to cleave off an extra, or precursor, sequence of RNA on tRNA...
. Then, according to this hypothesis, the seven ancestral eukaryote LSm genes duplicated again to seven pairs of Sm/LSm paralogs; LSm1/SmB, LSm2/SmD1, LSm3/SmD2, LSm4/SmD3, LSm5/SmE, LSm6/SmF and LSm7/SmG. These two group of seven LSm genes (and the corresponding two kinds of LSm rings) evolved to an Sm ring (requiring RNA) and a Lsm ring (which forms without RNA). The LSm1/LSm8 paralog pair also seems to have originated prior to the last common eukaryote ancestor, for a total of at least 15 LSm protein genes. The SmD1/LSm10 paralog pair and the SmD2/LSm11 paralog pair exist only in animal
Animal
Animals are a major group of multicellular, eukaryotic organisms of the kingdom Animalia or Metazoa. Their body plan eventually becomes fixed as they develop, although some undergo a process of metamorphosis later on in their life. Most animals are motile, meaning they can move spontaneously and...
s, fungi
Fungus
A fungus is a member of a large group of eukaryotic organisms that includes microorganisms such as yeasts and molds , as well as the more familiar mushrooms. These organisms are classified as a kingdom, Fungi, which is separate from plants, animals, and bacteria...
, and the amoebozoa
Amoebozoa
The Amoebozoa are a major group of amoeboid protozoa, including the majority that move by means ofinternal cytoplasmic flow. Their pseudopodia are characteristically blunt and finger-like,...
(sometimes identified as the unikont
Unikont
Unikonts are members of the Unikonta, a taxonomic group proposed by Thomas Cavalier-Smith.It includes amoebozoa, opisthokonts, and Apusozoa.-Clade:...
clade) and appears to be absent in the bikont
Bikont
A Bikont is a eukaryotic cell with two flagella, as its name suggests. It is a division of eukaryotes.-Enzymes:Another shared trait of bikonts is the fusion of two genes into a single unit: the genes for thymidylate synthase and dihydrofolate reductase encode a singleprotein with two...
clade (chromalveolate
Chromalveolate
Chromalveolata is a eukaryote supergroup first proposed by Thomas Cavalier-Smith as a refinement of his kingdom Chromista, which was first put forward in 1981. Chromalveolata was proposed to represent the result of a single secondary endosymbiosis between a line descending from a bikont and a red...
s, excavate
Excavate
The excavates are a major kingdom of unicellular eukaryotes, often known as Excavata. The phylogenetic category Excavata, proposed by Cavalier-Smith in 2002, contains a variety of free-living and symbiotic forms, and also includes some important parasites of humans.-Characteristics:Many excavates...
s, plant
Plant
Plants are living organisms belonging to the kingdom Plantae. Precise definitions of the kingdom vary, but as the term is used here, plants include familiar organisms such as trees, flowers, herbs, bushes, grasses, vines, ferns, mosses, and green algae. The group is also called green plants or...
s and rhizaria
Rhizaria
The Rhizaria are a species-rich supergroup of unicellular eukaryotes. This supergroup was proposed by Cavalier-Smith in 2002. They vary considerably in form, but for the most part they are amoeboids with filose, reticulose, or microtubule-supported pseudopods...
). Therefore, these two gene duplications predated this fundamental split in the eukaryote lineage. The SmB/SmN paralog pair is seen only in the placental mammals
Eutheria
Eutheria is a group of mammals consisting of placental mammals plus all extinct mammals that are more closely related to living placentals than to living marsupials . They are distinguished from noneutherians by various features of the feet, ankles, jaws and teeth...
, which dates this LSm gene duplication.
Biogenesis of snRNPs
Small nuclear ribonucleoproteins (snRNPs) assemble in a tightly orchestrated and regulated process that involves both the cell nucleusCell nucleus
In cell biology, the nucleus is a membrane-enclosed organelle found in eukaryotic cells. It contains most of the cell's genetic material, organized as multiple long linear DNA molecules in complex with a large variety of proteins, such as histones, to form chromosomes. The genes within these...
and cytoplasm
Cytoplasm
The cytoplasm is a small gel-like substance residing between the cell membrane holding all the cell's internal sub-structures , except for the nucleus. All the contents of the cells of prokaryote organisms are contained within the cytoplasm...
.

