
Phylogenetics
Encyclopedia
In biology
, phylogenetics (faɪlɵdʒɪˈnɛtɪks) is the study of evolution
ary relatedness among groups of organism
s (e.g. species
, population
s), which is discovered through molecular sequencing data and morphological data matrices. The term phylogenetics derives from the Greek terms phyle (φυλή) and phylon (φῦλον), denoting “tribe” and “race”; and the term genetikos (γενετικός), denoting “relative to birth”, from genesis (γένεσις) “birth”.
Taxonomy
, the classification, identification, and naming of organism
s, is richly informed by phylogenetics, but remains methodologically and logically distinct. The fields of phylogenetics and taxonomy overlap in the science of phylogenetic systematics — one methodology, cladism (also cladistics
) shared derived characters (synapomorphies) used to create ancestor-descendant trees (cladograms) and delimit taxa (clades). In biological systematics
as a whole, phylogenetic analyses have become essential in researching the evolutionary tree of life
.
. This may be visualized in a phylogenetic tree
.
The problem posed by phylogenetics is that genetic
data are only available for living taxa, and the fossil
records (osteometric data) contains less data and more-ambiguous morphological characters. A phylogenetic tree represents a hypothesis of the order in which evolutionary events are assumed to have occurred.
Cladistics
is the current method of choice to infer phylogenetic trees. The most commonly-used methods
to infer phylogenies include parsimony, maximum likelihood
, and MCMC
-based Bayesian inference
. Phenetics
, popular in the mid-20th century but now largely obsolete, uses distance matrix
-based methods to construct trees based on overall similarity, which is often assumed to approximate phylogenetic relationships. All methods depend upon an implicit or explicit mathematical model
describing the evolution of characters observed in the species included, and are usually used for molecular phylogeny
, wherein the characters are aligned nucleotide
or amino acid
sequences.
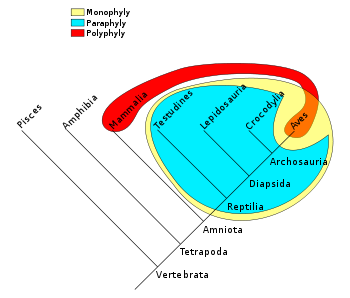 There are some terms that describe the nature of a grouping in such trees. For instance, all birds and reptiles are believed to have descended from a single common ancestor, so this taxonomic grouping (yellow in the diagram below) is called monophyletic
There are some terms that describe the nature of a grouping in such trees. For instance, all birds and reptiles are believed to have descended from a single common ancestor, so this taxonomic grouping (yellow in the diagram below) is called monophyletic
. "Modern reptile" (cyan
in the diagram) is a grouping that contains a common ancestor, but does not contain all descendants of that ancestor (birds are excluded). This is an example of a paraphyletic
group. A grouping such as warm-blooded
animals would include only mammals and birds (red/orange in the diagram) and is called polyphyletic
because the members of this grouping do not include the most recent common ancestor
.
s, often anatomical characteristics. Today, molecular data, which includes protein and DNA sequences, are used to construct phylogenetic trees.
The overall goal of National Science Foundation
's Assembling the Tree of Life activity (AToL) is to resolve evolutionary relationships for large groups of organisms throughout the history of life, with the research often involving large teams working across institutions and disciplines. Investigators are typically supported for projects in data acquisition, analysis, algorithm development and dissemination in computational phylogenetics and phyloinformatics. For example, RedToL
aims at reconstructing the Red Algal Tree of Life.
's recapitulation theory
, or biogenetic law, was widely accepted. This theory was often expressed as "ontogeny
recapitulates phylogeny", i.e. the development of an organism exactly mirrors the evolutionary development of the species. Haeckel's early version of this hypothesis [that the embryo mirrors adult evolutionary ancestors] has since been rejected, and the hypothesis amended as the embryo's development mirroring embryos of its evolutionary ancestors. He was accused by five professors of falsifying his images of embryos (See Ernst Haeckel
). Most modern biologists recognize numerous connections between ontogeny and phylogeny, explain them using evolutionary theory
, or view them as supporting evidence for that theory. Donald I. Williamson
suggested that larvae and embryos represented adults in other taxa that have been transferred by hybridization (the larval transfer theory). However, Williamson's views do not represent mainstream thought in molecular biology, and there is a significant body of evidence against the larval transfer theory.
. Vertical gene transfer is the passage of genes from parent to offspring, and horizontal gene transfer or lateral gene transfer occurs when genes jump between unrelated organisms, a common phenomenon in prokaryote
s; a good example of this is the acquired antibiotic resistance
as a result of gene exchange between some bacteria and development of multidrug resistant bacterial species.
Horizontal gene transfer has complicated the determination of phylogenies of organisms, and inconsistencies in phylogeny have been reported among specific groups of organisms depending on the genes used to construct evolutionary trees.
Carl Woese came up with the three-domain
theory of life (eubacteria, archaea
and eukaryota) based on his discovery that the genes encoding ribosomal RNA
are ancient and distributed over all lineages of life with little or no horizontal gene transfer. Therefore, rRNAs
are commonly recommended as molecular clocks for reconstructing phylogenies.
This has been particularly useful for the phylogeny of microorganisms, to which the species concept does not apply and which are too morphologically simple to be classified based on phenotypic traits.
, it has become feasible to gather large amounts of data (DNA or amino acid sequences) to infer phylogenetic hypotheses. For example, it is not rare to find studies with character matrices based on whole mitochondrial genomes (~16,000 nucleotides, in many animals). However, it has been proposed that it is more important to increase the number of taxa in the matrix than to increase the number of characters, because the more taxa the more robust is the resulting phylogenetic tree.
This may be partly due to the breaking up of long branches
. It has been argued that this is an important reason to incorporate data from fossils into phylogenies where possible. Of course, phylogenetic data that include fossil taxa are generally based on morphology, rather than DNA data. Using simulations, Derrick Zwickl and David Hillis
found that increasing taxon sampling in phylogenetic inference has a positive effect on the accuracy of phylogenetic analyses.
Another important factor that affects the accuracy of tree reconstruction is whether the data analyzed actually contain a useful phylogenetic signal, a term that is used generally to denote whether related organisms tend to resemble each other with respect to their genetic material or phenotypic traits. Ultimately, however, there is no way to measure whether a particular phylogenetic hypothesis is accurate or not, unless the "true" relationships among the taxa being examined are already known. The best result an empirical systematist can hope to attain is a tree with branches well-supported by the available evidence.
Molecular phylogenies can reveal rates of diversification, but in order to track rates of origination, extinction and patterns in diversification, fossil data must be incorporated. Molecular techniques assume a constant rate of diversification, which is rarely likely to be true; in some (but by no means all) cases, the assumptions inherent in interpreting the fossil record (e.g. a complete and unbiased record) are closer to being true than the assumption of a constant rate, making fossil insights more accurate than molecular reconstructions.
Biology
Biology is a natural science concerned with the study of life and living organisms, including their structure, function, growth, origin, evolution, distribution, and taxonomy. Biology is a vast subject containing many subdivisions, topics, and disciplines...
, phylogenetics (faɪlɵdʒɪˈnɛtɪks) is the study of evolution
Evolution
Evolution is any change across successive generations in the heritable characteristics of biological populations. Evolutionary processes give rise to diversity at every level of biological organisation, including species, individual organisms and molecules such as DNA and proteins.Life on Earth...
ary relatedness among groups of organism
Organism
In biology, an organism is any contiguous living system . In at least some form, all organisms are capable of response to stimuli, reproduction, growth and development, and maintenance of homoeostasis as a stable whole.An organism may either be unicellular or, as in the case of humans, comprise...
s (e.g. species
Species
In biology, a species is one of the basic units of biological classification and a taxonomic rank. A species is often defined as a group of organisms capable of interbreeding and producing fertile offspring. While in many cases this definition is adequate, more precise or differing measures are...
, population
Population
A population is all the organisms that both belong to the same group or species and live in the same geographical area. The area that is used to define a sexual population is such that inter-breeding is possible between any pair within the area and more probable than cross-breeding with individuals...
s), which is discovered through molecular sequencing data and morphological data matrices. The term phylogenetics derives from the Greek terms phyle (φυλή) and phylon (φῦλον), denoting “tribe” and “race”; and the term genetikos (γενετικός), denoting “relative to birth”, from genesis (γένεσις) “birth”.
Taxonomy
Alpha taxonomy
Alpha taxonomy is the discipline concerned with finding, describing and naming species of living or fossil organisms. This field is supported by institutions holding collections of these organisms, with relevant data, carefully curated: such institutes include natural history museums, herbaria and...
, the classification, identification, and naming of organism
Organism
In biology, an organism is any contiguous living system . In at least some form, all organisms are capable of response to stimuli, reproduction, growth and development, and maintenance of homoeostasis as a stable whole.An organism may either be unicellular or, as in the case of humans, comprise...
s, is richly informed by phylogenetics, but remains methodologically and logically distinct. The fields of phylogenetics and taxonomy overlap in the science of phylogenetic systematics — one methodology, cladism (also cladistics
Cladistics
Cladistics is a method of classifying species of organisms into groups called clades, which consist of an ancestor organism and all its descendants . For example, birds, dinosaurs, crocodiles, and all descendants of their most recent common ancestor form a clade...
) shared derived characters (synapomorphies) used to create ancestor-descendant trees (cladograms) and delimit taxa (clades). In biological systematics
Systematics
Biological systematics is the study of the diversification of terrestrial life, both past and present, and the relationships among living things through time. Relationships are visualized as evolutionary trees...
as a whole, phylogenetic analyses have become essential in researching the evolutionary tree of life
Tree of life (science)
Charles Darwin proposed that phylogeny, the evolutionary relatedness among species through time, was expressible as a metaphor he termed the Tree of Life...
.
Construction of a phylogenetic tree
Evolution is regarded as a branching process, whereby populations are altered over time and may speciate into separate branches, hybridize together, or terminate by extinctionExtinction
In biology and ecology, extinction is the end of an organism or of a group of organisms , normally a species. The moment of extinction is generally considered to be the death of the last individual of the species, although the capacity to breed and recover may have been lost before this point...
. This may be visualized in a phylogenetic tree
Phylogenetic tree
A phylogenetic tree or evolutionary tree is a branching diagram or "tree" showing the inferred evolutionary relationships among various biological species or other entities based upon similarities and differences in their physical and/or genetic characteristics...
.
The problem posed by phylogenetics is that genetic
Genetics
Genetics , a discipline of biology, is the science of genes, heredity, and variation in living organisms....
data are only available for living taxa, and the fossil
Fossil
Fossils are the preserved remains or traces of animals , plants, and other organisms from the remote past...
records (osteometric data) contains less data and more-ambiguous morphological characters. A phylogenetic tree represents a hypothesis of the order in which evolutionary events are assumed to have occurred.
Cladistics
Cladistics
Cladistics is a method of classifying species of organisms into groups called clades, which consist of an ancestor organism and all its descendants . For example, birds, dinosaurs, crocodiles, and all descendants of their most recent common ancestor form a clade...
is the current method of choice to infer phylogenetic trees. The most commonly-used methods
Computational phylogenetics
Computational phylogenetics is the application of computational algorithms, methods and programs to phylogenetic analyses. The goal is to assemble a phylogenetic tree representing a hypothesis about the evolutionary ancestry of a set of genes, species, or other taxa...
to infer phylogenies include parsimony, maximum likelihood
Maximum likelihood
In statistics, maximum-likelihood estimation is a method of estimating the parameters of a statistical model. When applied to a data set and given a statistical model, maximum-likelihood estimation provides estimates for the model's parameters....
, and MCMC
Markov chain Monte Carlo
Markov chain Monte Carlo methods are a class of algorithms for sampling from probability distributions based on constructing a Markov chain that has the desired distribution as its equilibrium distribution. The state of the chain after a large number of steps is then used as a sample of the...
-based Bayesian inference
Bayesian inference
In statistics, Bayesian inference is a method of statistical inference. It is often used in science and engineering to determine model parameters, make predictions about unknown variables, and to perform model selection...
. Phenetics
Phenetics
In biology, phenetics, also known as taximetrics, is an attempt to classify organisms based on overall similarity, usually in morphology or other observable traits, regardless of their phylogeny or evolutionary relation. It is closely related to numerical taxonomy which is concerned with the use of...
, popular in the mid-20th century but now largely obsolete, uses distance matrix
Distance matrix
In mathematics, computer science and graph theory, a distance matrix is a matrix containing the distances, taken pairwise, of a set of points...
-based methods to construct trees based on overall similarity, which is often assumed to approximate phylogenetic relationships. All methods depend upon an implicit or explicit mathematical model
Mathematical model
A mathematical model is a description of a system using mathematical concepts and language. The process of developing a mathematical model is termed mathematical modeling. Mathematical models are used not only in the natural sciences and engineering disciplines A mathematical model is a...
describing the evolution of characters observed in the species included, and are usually used for molecular phylogeny
Molecular phylogeny
Molecular phylogenetics is the analysis of hereditary molecular differences, mainly in DNA sequences, to gain information on an organism's evolutionary relationships. The result of a molecular phylogenetic analysis is expressed in a phylogenetic tree...
, wherein the characters are aligned nucleotide
Nucleotide
Nucleotides are molecules that, when joined together, make up the structural units of RNA and DNA. In addition, nucleotides participate in cellular signaling , and are incorporated into important cofactors of enzymatic reactions...
or amino acid
Amino acid
Amino acids are molecules containing an amine group, a carboxylic acid group and a side-chain that varies between different amino acids. The key elements of an amino acid are carbon, hydrogen, oxygen, and nitrogen...
sequences.
Grouping of organisms

Monophyly
In common cladistic usage, a monophyletic group is a taxon which forms a clade, meaning that it contains all the descendants of the possibly hypothetical closest common ancestor of the members of the group. The term is synonymous with the uncommon term holophyly...
. "Modern reptile" (cyan
Cyan
Cyan from , transliterated: kýanos, meaning "dark blue substance") may be used as the name of any of a number of colors in the blue/green range of the spectrum. In reference to the visible spectrum cyan is used to refer to the color obtained by mixing equal amounts of green and blue light or the...
in the diagram) is a grouping that contains a common ancestor, but does not contain all descendants of that ancestor (birds are excluded). This is an example of a paraphyletic
Paraphyly
A group of taxa is said to be paraphyletic if the group consists of all the descendants of a hypothetical closest common ancestor minus one or more monophyletic groups of descendants...
group. A grouping such as warm-blooded
Warm-blooded
The term warm-blooded is a colloquial term to describe animal species which have a relatively higher blood temperature, and maintain thermal homeostasis primarily through internal metabolic processes...
animals would include only mammals and birds (red/orange in the diagram) and is called polyphyletic
Polyphyly
A polyphyletic group is one whose members' last common ancestor is not a member of the group.For example, the group consisting of warm-blooded animals is polyphyletic, because it contains both mammals and birds, but the most recent common ancestor of mammals and birds was cold-blooded...
because the members of this grouping do not include the most recent common ancestor
Most recent common ancestor
In genetics, the most recent common ancestor of any set of organisms is the most recent individual from which all organisms in the group are directly descended...
.
Molecular phylogenetics
The evolutionary connections between organisms are represented graphically through phylogenetic trees. Due to the fact that evolution takes place over long periods of time that cannot be observed directly, biologists must reconstruct phylogenies by inferring the evolutionary relationships among present-day organisms. Fossils can aid with the reconstruction of phylogenies; however, fossil records are often too poor to be of good help. Therefore, biologists tend to be restricted with analysing present-day organisms to identify their evolutionary relationships. Phylogenetic relationships in the past were reconstructed by looking at phenotypePhenotype
A phenotype is an organism's observable characteristics or traits: such as its morphology, development, biochemical or physiological properties, behavior, and products of behavior...
s, often anatomical characteristics. Today, molecular data, which includes protein and DNA sequences, are used to construct phylogenetic trees.
The overall goal of National Science Foundation
National Science Foundation
The National Science Foundation is a United States government agency that supports fundamental research and education in all the non-medical fields of science and engineering. Its medical counterpart is the National Institutes of Health...
's Assembling the Tree of Life activity (AToL) is to resolve evolutionary relationships for large groups of organisms throughout the history of life, with the research often involving large teams working across institutions and disciplines. Investigators are typically supported for projects in data acquisition, analysis, algorithm development and dissemination in computational phylogenetics and phyloinformatics. For example, RedToL
RedToL
RedToL, or Red Algal Tree of Life, is part of the collaborative National Science Foundation Assembling the Tree of Life activity , funded through the Division of Environmental Biology, Directorate for Biological Sciences...
aims at reconstructing the Red Algal Tree of Life.
Ernst Haeckel's recapitulation theory
During the late 19th century, Ernst HaeckelErnst Haeckel
The "European War" became known as "The Great War", and it was not until 1920, in the book "The First World War 1914-1918" by Charles à Court Repington, that the term "First World War" was used as the official name for the conflict.-Research:...
's recapitulation theory
Recapitulation theory
The theory of recapitulation, also called the biogenetic law or embryological parallelism—and often expressed as "ontogeny recapitulates phylogeny"—is a disproven hypothesis that in developing from embryo to adult, animals go through stages resembling or representing successive stages...
, or biogenetic law, was widely accepted. This theory was often expressed as "ontogeny
Ontogeny
Ontogeny is the origin and the development of an organism – for example: from the fertilized egg to mature form. It covers in essence, the study of an organism's lifespan...
recapitulates phylogeny", i.e. the development of an organism exactly mirrors the evolutionary development of the species. Haeckel's early version of this hypothesis [that the embryo mirrors adult evolutionary ancestors] has since been rejected, and the hypothesis amended as the embryo's development mirroring embryos of its evolutionary ancestors. He was accused by five professors of falsifying his images of embryos (See Ernst Haeckel
Ernst Haeckel
The "European War" became known as "The Great War", and it was not until 1920, in the book "The First World War 1914-1918" by Charles à Court Repington, that the term "First World War" was used as the official name for the conflict.-Research:...
). Most modern biologists recognize numerous connections between ontogeny and phylogeny, explain them using evolutionary theory
Evolutionary developmental biology
Evolutionary developmental biology is a field of biology that compares the developmental processes of different organisms to determine the ancestral relationship between them, and to discover how developmental processes evolved...
, or view them as supporting evidence for that theory. Donald I. Williamson
Donald I. Williamson
Donald Irving Williamson is a British planktologist and carcinologist, born 8 January 1922, Alnham, Northumberland, England. He gained his first degree from the Durham University in 1942, his Ph.D. from the same university in 1948, and a D.Sc. from the Newcastle University in 1972...
suggested that larvae and embryos represented adults in other taxa that have been transferred by hybridization (the larval transfer theory). However, Williamson's views do not represent mainstream thought in molecular biology, and there is a significant body of evidence against the larval transfer theory.
Gene transfer
In general, organisms can inherit genes in two ways: vertical gene transfer and horizontal gene transferHorizontal gene transfer
Horizontal gene transfer , also lateral gene transfer , is any process in which an organism incorporates genetic material from another organism without being the offspring of that organism...
. Vertical gene transfer is the passage of genes from parent to offspring, and horizontal gene transfer or lateral gene transfer occurs when genes jump between unrelated organisms, a common phenomenon in prokaryote
Prokaryote
The prokaryotes are a group of organisms that lack a cell nucleus , or any other membrane-bound organelles. The organisms that have a cell nucleus are called eukaryotes. Most prokaryotes are unicellular, but a few such as myxobacteria have multicellular stages in their life cycles...
s; a good example of this is the acquired antibiotic resistance
Antibiotic resistance
Antibiotic resistance is a type of drug resistance where a microorganism is able to survive exposure to an antibiotic. While a spontaneous or induced genetic mutation in bacteria may confer resistance to antimicrobial drugs, genes that confer resistance can be transferred between bacteria in a...
as a result of gene exchange between some bacteria and development of multidrug resistant bacterial species.
Horizontal gene transfer has complicated the determination of phylogenies of organisms, and inconsistencies in phylogeny have been reported among specific groups of organisms depending on the genes used to construct evolutionary trees.
Carl Woese came up with the three-domain
Domain (biology)
In biological taxonomy, a domain is the highest taxonomic rank of organisms, higher than a kingdom. According to the three-domain system of Carl Woese, introduced in 1990, the Tree of Life consists of three domains: Archaea, Bacteria and Eukarya...
theory of life (eubacteria, archaea
Archaea
The Archaea are a group of single-celled microorganisms. A single individual or species from this domain is called an archaeon...
and eukaryota) based on his discovery that the genes encoding ribosomal RNA
Ribosomal RNA
Ribosomal ribonucleic acid is the RNA component of the ribosome, the enzyme that is the site of protein synthesis in all living cells. Ribosomal RNA provides a mechanism for decoding mRNA into amino acids and interacts with tRNAs during translation by providing peptidyl transferase activity...
are ancient and distributed over all lineages of life with little or no horizontal gene transfer. Therefore, rRNAs
RNA
Ribonucleic acid , or RNA, is one of the three major macromolecules that are essential for all known forms of life....
are commonly recommended as molecular clocks for reconstructing phylogenies.
This has been particularly useful for the phylogeny of microorganisms, to which the species concept does not apply and which are too morphologically simple to be classified based on phenotypic traits.
Taxon sampling and phylogenetic signal
Owing to the development of advanced sequencing techniques in molecular biologyMolecular biology
Molecular biology is the branch of biology that deals with the molecular basis of biological activity. This field overlaps with other areas of biology and chemistry, particularly genetics and biochemistry...
, it has become feasible to gather large amounts of data (DNA or amino acid sequences) to infer phylogenetic hypotheses. For example, it is not rare to find studies with character matrices based on whole mitochondrial genomes (~16,000 nucleotides, in many animals). However, it has been proposed that it is more important to increase the number of taxa in the matrix than to increase the number of characters, because the more taxa the more robust is the resulting phylogenetic tree.
This may be partly due to the breaking up of long branches
Long branch attraction
Long branch attraction is a phenomenon in phylogenetic analyses when rapidly evolving lineages are inferred to be closely related, regardless of their true evolutionary relationships. For example, in DNA sequence-based analyses, the problem arises when sequences from two lineages evolve rapidly...
. It has been argued that this is an important reason to incorporate data from fossils into phylogenies where possible. Of course, phylogenetic data that include fossil taxa are generally based on morphology, rather than DNA data. Using simulations, Derrick Zwickl and David Hillis
David Hillis
David Mark Hillis is an American evolutionary biologist, and the Alfred W. Roark Centennial Professor of Biology at the University of Texas at Austin. He is best known for his studies of molecular evolution, phylogeny, and vertebrate systematics...
found that increasing taxon sampling in phylogenetic inference has a positive effect on the accuracy of phylogenetic analyses.
Another important factor that affects the accuracy of tree reconstruction is whether the data analyzed actually contain a useful phylogenetic signal, a term that is used generally to denote whether related organisms tend to resemble each other with respect to their genetic material or phenotypic traits. Ultimately, however, there is no way to measure whether a particular phylogenetic hypothesis is accurate or not, unless the "true" relationships among the taxa being examined are already known. The best result an empirical systematist can hope to attain is a tree with branches well-supported by the available evidence.
Importance of missing data
In general, the more data that is available when constructing a tree, the more accurate and reliable the resulting tree will be. Missing data is no less detrimental than simply having less data, although its impact is greatest when most of the missing data is in a small number of taxa. The fewer characters that have missing data, the better; concentrating the missing data across a small number of character states produces a more robust tree.Role of fossils
Because many morphological characters involve embryological or soft-tissue characters that cannot be fossilized, and the interpretation of fossils is more ambiguous than living taxa, it is sometimes difficult to incorporate fossil data into phylogenies. However, despite these limitations, the inclusion of fossils is invaluable, as they can provide information in sparse areas of trees, breaking up long branches and constraining intermediate character states; thus, fossil taxa contribute as much to tree resolution as modern taxa.Molecular phylogenies can reveal rates of diversification, but in order to track rates of origination, extinction and patterns in diversification, fossil data must be incorporated. Molecular techniques assume a constant rate of diversification, which is rarely likely to be true; in some (but by no means all) cases, the assumptions inherent in interpreting the fossil record (e.g. a complete and unbiased record) are closer to being true than the assumption of a constant rate, making fossil insights more accurate than molecular reconstructions.
Homoplasy weighting
Certain characters are more likely to be evolved convergently than others; logically, such characters should be given less weight in the reconstruction of a tree. Unfortunately the only objective way to determine convergence is by the construction of a tree – a somewhat circular method. Even so, weighting homoplasious characters does indeed lead to better-supported trees. Further refinement can be brought by weighting changes in one direction higher than changes in another; for instance, the presence of thoracic wings almost guarantees placement among the pterygote insects, although because wings are often lost secondarily, their absence does not exclude a taxon from the group.Further reading
- Schuh, R. T. and A. V. Z. Brower. 2009. Biological Systematics: principles and applications (2nd edn.) ISBN 978-0-8014-4799-0
External links
- The Tree of Life
- Interactive Tree of Life
- PhyloCode
- ExploreTree
- UCMP Exhibit Halls: Phylogeny Wing
- Willi Hennig Society
- Filogenetica.org in Spanish
- PhyloPat, Phylogenetic Patterns
- SplitsTree, program for computing phylogenetic trees and unrooted phylogenetic networks
- Dendroscope, program for drawing phylogenetic trees and rooted phylogenetic networks
- Phylogenetic inferring on the T-REX server
- Mesquite
- NCBI – Systematics and Molecular Phylogenetics
- What Genomes Can Tell Us About the Past – lecture on phylogenetics by Sydney BrennerSydney BrennerSydney Brenner, CH FRS is a South African biologist and a 2002 Nobel prize in Physiology or Medicine laureate, shared with H...
- Mikko's Phylogeny Archive
- Phylogenetic Reconstruction from Gene-Order Data
- ETE: A Python Environment for Tree Exploration This is a programming library to analyze, manipulate and visualize phylogenetic trees. Ref.
- PhylomeDBPhylomeDBPhylomeDB is a database of complete phylomes derived for different genomes within a specific taxonomic range....
: A public database hosting thousands of gene phylogenies ranging many different species.Ref.
- http://lifescied.org/content/9/4/513.fullA published teaching exercising for demonstrating the process of molecular phylogenetics

