Spliceosome
Encyclopedia
A spliceosome is a complex of snRNA and protein
subunits that removes intron
s from a transcribed
pre-mRNA
(hnRNA) segment. This process is generally referred to as splicing
.
s (snRNAs), and a range of associated protein factors.
The snRNAs that make up the nuclear
spliceosome are named U1, U2, U4, U5, and U6, and participate in several RNA-RNA and RNA-protein interactions. The RNA component of the small nuclear ribonucleic protein or snRNP is rich in uridine
(the nucleoside
analog of the uracil
nucleotide
).
The canonical assembly of the spliceosome occurs anew on each hnRNA (pre-mRNA). The hnRNA contains specific sequence elements that are recognized and utilized during spliceosome assembly. These include the 5' end splice, the branch point sequence, the polypyrimidine tract, and the 3' end splice site. The spliceosome catalyzes the removal of introns, and the ligation of the flanking exons.
Introns typically have a GU nucleotide sequence at the 5' end splice site, and an AG at the 3' end splice site. The 3' splice site can be further defined by a variable length of polypyrimidines, called the polypyrimidine tract
(PPT), which serves the dual function of recruiting factors to the 3' splice site and possibly recruiting factors to the branch point sequence (BPS). The BPS contains the conserved Adenosine required for the first step of splicing.
A group of less abundant snRNAs, U11, U12, U4atac
, and U6atac
, together with U5, are subunits of the so-called minor spliceosome
that splices a rare class of pre-mRNA introns, denoted U12-type. These snRNPs form the U12 spliceosome are located in the cytosol
.
New evidence derived from the first crystal structure of a group II intron suggests that the spliceosome is actually a ribozyme
, and that it uses a two–metal ion mechanism for catalysis.
s) is a major source of genetic diversity
in eukaryotes. Splice variants have been used to account for the relatively small number of gene
s in the human genome
. For years the estimate widely varied with top estimates reaching 100,000 genes, but now, due to the Human Genome Project
the figure is believed to be closer to 20,000 genes. One particular Drosophila
gene (Dscam
, the Drosophila homolog of the human Down syndrome cell adhesion molecule DSCAM) can be alternatively spliced into 38,000 different mRNA.
and Richard J. Roberts
were awarded the 1993 Nobel Prize in Physiology or Medicine
for their discovery of introns and the splicing process.
(U2 snRNP auxiliary factor) and possibly U1 snRNP. In an ATP-dependent reaction, U2 snRNP becomes tightly associated with the branch point sequence (BPS) to form complex A. A duplex formed between U2 snRNP and the hnRNA branch region bulges out the branch adenosine specifying it as the nucleophile for the first transesterification.
The presence of a pseudouridine residue in U2 snRNA, nearly opposite of the branch site, results in an altered conformation of the RNA-RNA duplex upon the U2 snRNP binding. Specifically, the altered structure of the duplex induced by the pseudouridine places the 2' OH of the bulged adenosine in a favorable position for the first step of splicing. The U4/U5/U6 tri-snRNP (see Figure 1) is recruited to the assembling spliceosome to form complex B, and following several rearrangements, complex C (the spliceosome) is activated for catalysis. It is unclear how the triple snRNP is recruited to complex A, but this process may be mediated through protein-protein interactions and/or base pairing interactions between U2 snRNA and U6 snRNA.
The U5 snRNP interacts with sequences at the 5' and 3' splice sites via the invariant loop of U5 snRNA and U5 protein components interact with the 3' splice site region.
Upon recruitment of the triple snRNP, several RNA-RNA rearrangements precede the first catalytic step and further rearrangements occur in the catalytically active spliceosome. Several of the RNA-RNA interactions are mutually exclusive; however, it is not known what triggers these interactions, nor the order of these rearrangements. The first rearrangement is probably the displacement of U1 snRNP from the 5' splice site and formation of a U6 snRNA interaction. It is known that U1 snRNP is only weakly associated with fully formed spliceosomes, and U1 snRNP is inhibitory to the formation of a U6-5' splice site interaction on a model of substrate oligonucleotide containing a short 5' exon and 5' splice site. Binding of U2 snRNP to the branch point sequence (BPS) is one example of an RNA-RNA interaction displacing a protein-RNA interaction. Upon recruitment of U2 snRNP, the branch binding protein SF1 in the commitment complex is displaced since the binding site of U2 snRNA and SF1 are mutually exclusive events.
Within the U2 snRNA, there are other mutually exclusive rearrangements that occur between competing conformations. For example, in the active form, stem loop IIa is favored; in the inactive form a mutually exclusive interaction between the loop and a downstream sequence predominates. It is unclear how U4 is displaced from U6 snRNAm, although RNA has been implicated in spliceosome assembly, and may function to unwind U4/U6 and promote the formation of a U2/U6 snRNA interaction. The interactions of U4/U6 stem loops I and II dissociate and the freed stem loop II region of U6 folds on itself to form an intramolecular stem loop and U4 is no longer required in further spliceosome assembly. The freed stem loop I region of U6 base pairs with U2 snRNA forming the U2/U6 helix I. However, the helix I structure is mutually exclusive with the 3' half of an internal 5' stem loop region of U2 snRNA.
Protein
Proteins are biochemical compounds consisting of one or more polypeptides typically folded into a globular or fibrous form, facilitating a biological function. A polypeptide is a single linear polymer chain of amino acids bonded together by peptide bonds between the carboxyl and amino groups of...
subunits that removes intron
Intron
An intron is any nucleotide sequence within a gene that is removed by RNA splicing to generate the final mature RNA product of a gene. The term intron refers to both the DNA sequence within a gene, and the corresponding sequence in RNA transcripts. Sequences that are joined together in the final...
s from a transcribed
Transcription (genetics)
Transcription is the process of creating a complementary RNA copy of a sequence of DNA. Both RNA and DNA are nucleic acids, which use base pairs of nucleotides as a complementary language that can be converted back and forth from DNA to RNA by the action of the correct enzymes...
pre-mRNA
Messenger RNA
Messenger RNA is a molecule of RNA encoding a chemical "blueprint" for a protein product. mRNA is transcribed from a DNA template, and carries coding information to the sites of protein synthesis: the ribosomes. Here, the nucleic acid polymer is translated into a polymer of amino acids: a protein...
(hnRNA) segment. This process is generally referred to as splicing
Splicing (genetics)
In molecular biology and genetics, splicing is a modification of an RNA after transcription, in which introns are removed and exons are joined. This is needed for the typical eukaryotic messenger RNA before it can be used to produce a correct protein through translation...
.
Composition
Each spliceosome is composed of five small nuclear RNASmall nuclear RNA
Small nuclear ribonucleic acid is a class of small RNA molecules that are found within the nucleus of eukaryotic cells. They are transcribed by RNA polymerase II or RNA polymerase III and are involved in a variety of important processes such as RNA splicing , regulation of transcription factors ...
s (snRNAs), and a range of associated protein factors.
The snRNAs that make up the nuclear
Cell nucleus
In cell biology, the nucleus is a membrane-enclosed organelle found in eukaryotic cells. It contains most of the cell's genetic material, organized as multiple long linear DNA molecules in complex with a large variety of proteins, such as histones, to form chromosomes. The genes within these...
spliceosome are named U1, U2, U4, U5, and U6, and participate in several RNA-RNA and RNA-protein interactions. The RNA component of the small nuclear ribonucleic protein or snRNP is rich in uridine
Uridine
Uridine is a molecule that is formed when uracil is attached to a ribose ring via a β-N1-glycosidic bond.If uracil is attached to a deoxyribose ring, it is known as a deoxyuridine....
(the nucleoside
Nucleoside
Nucleosides are glycosylamines consisting of a nucleobase bound to a ribose or deoxyribose sugar via a beta-glycosidic linkage...
analog of the uracil
Uracil
Uracil is one of the four nucleobases in the nucleic acid of RNA that are represented by the letters A, G, C and U. The others are adenine, cytosine, and guanine. In RNA, uracil binds to adenine via two hydrogen bonds. In DNA, the uracil nucleobase is replaced by thymine.Uracil is a common and...
nucleotide
Nucleotide
Nucleotides are molecules that, when joined together, make up the structural units of RNA and DNA. In addition, nucleotides participate in cellular signaling , and are incorporated into important cofactors of enzymatic reactions...
).
The canonical assembly of the spliceosome occurs anew on each hnRNA (pre-mRNA). The hnRNA contains specific sequence elements that are recognized and utilized during spliceosome assembly. These include the 5' end splice, the branch point sequence, the polypyrimidine tract, and the 3' end splice site. The spliceosome catalyzes the removal of introns, and the ligation of the flanking exons.
Introns typically have a GU nucleotide sequence at the 5' end splice site, and an AG at the 3' end splice site. The 3' splice site can be further defined by a variable length of polypyrimidines, called the polypyrimidine tract
Polypyrimidine tract
The polypyrimidine tract is a region of messenger RNA that promotes the assembly of the spliceosome, the protein complex specialized for carrying out RNA splicing during the process of post-transcriptional modification...
(PPT), which serves the dual function of recruiting factors to the 3' splice site and possibly recruiting factors to the branch point sequence (BPS). The BPS contains the conserved Adenosine required for the first step of splicing.
A group of less abundant snRNAs, U11, U12, U4atac
U4atac minor spliceosomal RNA
U4atac minor spliceosomal RNA is a ncRNA which is an essential component of the minor U12-type spliceosome complex. The U12-type spliceosome is required for removal of the rarer class of eukaryotic introns ....
, and U6atac
U6atac minor spliceosomal RNA
U6atac minor spliceosomal RNA is a non-coding RNA which is an essential component of the minor U12-type spliceosome complex. The U12-type spliceosome is required for removal of the rarer class of eukaryotic introns ....
, together with U5, are subunits of the so-called minor spliceosome
Minor spliceosome
The minor spliceosome is a ribonucleoprotein complex that catalyses the removal of an atypical class of spliceosomal introns from eukaryotic messenger RNAs in plant, insects, vertebrates and some fungi . This process is called noncanonical splicing, as opposed to U2-dependent canonical splicing...
that splices a rare class of pre-mRNA introns, denoted U12-type. These snRNPs form the U12 spliceosome are located in the cytosol
Cytosol
The cytosol or intracellular fluid is the liquid found inside cells, that is separated into compartments by membranes. For example, the mitochondrial matrix separates the mitochondrion into compartments....
.
New evidence derived from the first crystal structure of a group II intron suggests that the spliceosome is actually a ribozyme
Ribozyme
A ribozyme is an RNA molecule with a well defined tertiary structure that enables it to catalyze a chemical reaction. Ribozyme means ribonucleic acid enzyme. It may also be called an RNA enzyme or catalytic RNA. Many natural ribozymes catalyze either the hydrolysis of one of their own...
, and that it uses a two–metal ion mechanism for catalysis.

Alternative splicing
Alternative splicing (the re-combination of different exonExon
An exon is a nucleic acid sequence that is represented in the mature form of an RNA molecule either after portions of a precursor RNA have been removed by cis-splicing or when two or more precursor RNA molecules have been ligated by trans-splicing. The mature RNA molecule can be a messenger RNA...
s) is a major source of genetic diversity
Genetic diversity
Genetic diversity, the level of biodiversity, refers to the total number of genetic characteristics in the genetic makeup of a species. It is distinguished from genetic variability, which describes the tendency of genetic characteristics to vary....
in eukaryotes. Splice variants have been used to account for the relatively small number of gene
Gene
A gene is a molecular unit of heredity of a living organism. It is a name given to some stretches of DNA and RNA that code for a type of protein or for an RNA chain that has a function in the organism. Living beings depend on genes, as they specify all proteins and functional RNA chains...
s in the human genome
Human genome
The human genome is the genome of Homo sapiens, which is stored on 23 chromosome pairs plus the small mitochondrial DNA. 22 of the 23 chromosomes are autosomal chromosome pairs, while the remaining pair is sex-determining...
. For years the estimate widely varied with top estimates reaching 100,000 genes, but now, due to the Human Genome Project
Human Genome Project
The Human Genome Project is an international scientific research project with a primary goal of determining the sequence of chemical base pairs which make up DNA, and of identifying and mapping the approximately 20,000–25,000 genes of the human genome from both a physical and functional...
the figure is believed to be closer to 20,000 genes. One particular Drosophila
Drosophila
Drosophila is a genus of small flies, belonging to the family Drosophilidae, whose members are often called "fruit flies" or more appropriately pomace flies, vinegar flies, or wine flies, a reference to the characteristic of many species to linger around overripe or rotting fruit...
gene (Dscam
DSCAM
DSCAM and Dscam are both abbreviations for Down Syndrome Cell Adhesion Molecule. The case difference is significant: DSCAM refers to the human protein, and Dscam refers to its analog in Drosophila....
, the Drosophila homolog of the human Down syndrome cell adhesion molecule DSCAM) can be alternatively spliced into 38,000 different mRNA.
RNA splicing
In 1977, work by the Sharp and Roberts labs revealed that genes of higher organisms are "split" or present in several distinct segments along the DNA molecule. The coding regions of the gene are separated by non-coding DNA that is not involved in protein expression. The split gene structure was found when adenoviral mRNAs were hybridized to endonuclease cleavage fragments of single stranded viral DNA. It was observed that the mRNAs of the mRNA-DNA hybrids contained 5' and 3' tails of non-hydrogen bonded regions. When larger fragments of viral DNAs were used, forked structures of looped out DNA were observed when hybridized to the viral mRNAs. It was realized that the looped out regions, the introns, are excised from the precursor mRNAs in a process Sharp named "splicing". The split gene structure was subsequently found to be common to most eukaryotic genes. Phillip SharpPhillip Allen Sharp
Phillip Allen Sharp is an American geneticist and molecular biologist who co-discovered RNA splicing. He shared the 1993 Nobel Prize in Physiology or Medicine with Richard J...
and Richard J. Roberts
Richard J. Roberts
Sir Richard "Rich" John Roberts is a British biochemist and molecular biologist. He was awarded the 1993 Nobel Prize in Physiology or Medicine with Phillip Allen Sharp for the discovery of introns in eukaryotic DNA and the mechanism of gene-splicing.When he was 4, his family moved to Bath. In...
were awarded the 1993 Nobel Prize in Physiology or Medicine
Nobel Prize in Physiology or Medicine
The Nobel Prize in Physiology or Medicine administered by the Nobel Foundation, is awarded once a year for outstanding discoveries in the field of life science and medicine. It is one of five Nobel Prizes established in 1895 by Swedish chemist Alfred Nobel, the inventor of dynamite, in his will...
for their discovery of introns and the splicing process.
Spliceosome assembly
The model for formation of the spliceosome active site involves an ordered, stepwise assembly of discrete snRNP particles on the hnRNA substrate. The first recognition of hnRNAs involves U1 snRNP binding to the 5' end splice site of the hnRNA and other non-snRNP associated factors to form the commitment complex, or early (E) complex in mammals. The commitment complex is an ATP-independent complex that commits the hnRNA to the splicing pathway. U2 snRNP is recruited to the branch region through interactions with the E complex component U2AFU2AF2
Splicing factor U2AF 65 kDa subunit is a protein that in humans is encoded by the U2AF2 gene.-Interactions:U2AF2 has been shown to interact with WT1, PUF60, SFRS2IP, SRPK2, SF1, U2 small nuclear RNA auxiliary factor 1 and SFRS11.-References:...
(U2 snRNP auxiliary factor) and possibly U1 snRNP. In an ATP-dependent reaction, U2 snRNP becomes tightly associated with the branch point sequence (BPS) to form complex A. A duplex formed between U2 snRNP and the hnRNA branch region bulges out the branch adenosine specifying it as the nucleophile for the first transesterification.
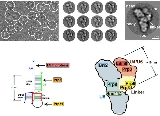
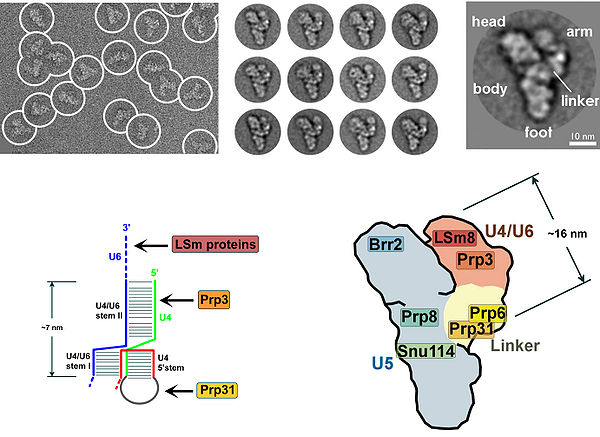
The presence of a pseudouridine residue in U2 snRNA, nearly opposite of the branch site, results in an altered conformation of the RNA-RNA duplex upon the U2 snRNP binding. Specifically, the altered structure of the duplex induced by the pseudouridine places the 2' OH of the bulged adenosine in a favorable position for the first step of splicing. The U4/U5/U6 tri-snRNP (see Figure 1) is recruited to the assembling spliceosome to form complex B, and following several rearrangements, complex C (the spliceosome) is activated for catalysis. It is unclear how the triple snRNP is recruited to complex A, but this process may be mediated through protein-protein interactions and/or base pairing interactions between U2 snRNA and U6 snRNA.
The U5 snRNP interacts with sequences at the 5' and 3' splice sites via the invariant loop of U5 snRNA and U5 protein components interact with the 3' splice site region.
Upon recruitment of the triple snRNP, several RNA-RNA rearrangements precede the first catalytic step and further rearrangements occur in the catalytically active spliceosome. Several of the RNA-RNA interactions are mutually exclusive; however, it is not known what triggers these interactions, nor the order of these rearrangements. The first rearrangement is probably the displacement of U1 snRNP from the 5' splice site and formation of a U6 snRNA interaction. It is known that U1 snRNP is only weakly associated with fully formed spliceosomes, and U1 snRNP is inhibitory to the formation of a U6-5' splice site interaction on a model of substrate oligonucleotide containing a short 5' exon and 5' splice site. Binding of U2 snRNP to the branch point sequence (BPS) is one example of an RNA-RNA interaction displacing a protein-RNA interaction. Upon recruitment of U2 snRNP, the branch binding protein SF1 in the commitment complex is displaced since the binding site of U2 snRNA and SF1 are mutually exclusive events.
Within the U2 snRNA, there are other mutually exclusive rearrangements that occur between competing conformations. For example, in the active form, stem loop IIa is favored; in the inactive form a mutually exclusive interaction between the loop and a downstream sequence predominates. It is unclear how U4 is displaced from U6 snRNAm, although RNA has been implicated in spliceosome assembly, and may function to unwind U4/U6 and promote the formation of a U2/U6 snRNA interaction. The interactions of U4/U6 stem loops I and II dissociate and the freed stem loop II region of U6 folds on itself to form an intramolecular stem loop and U4 is no longer required in further spliceosome assembly. The freed stem loop I region of U6 base pairs with U2 snRNA forming the U2/U6 helix I. However, the helix I structure is mutually exclusive with the 3' half of an internal 5' stem loop region of U2 snRNA.

