
Minor spliceosome
Encyclopedia
The minor spliceosome is a ribonucleoprotein
complex that catalyses the removal (splicing
) of an atypical class of spliceosomal introns (U12-type) from eukaryotic messenger RNA
s in plant, insects, vertebrates and some fungi (Rhizopus oryzae
). This process is called noncanonical splicing, as opposed to U2-dependent canonical splicing. U12-type introns represent less than 1% of all introns in human cells. However they are found in genes performing essential cellular functions.
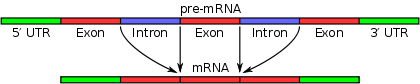
 A notable feature of eukaryotic nuclear pre mRNA introns is the relatively high level of conservation of the primary sequences of 5’ and 3’ splice sites over a great range of organisms.
A notable feature of eukaryotic nuclear pre mRNA introns is the relatively high level of conservation of the primary sequences of 5’ and 3’ splice sites over a great range of organisms.
Since 1989 till 1991, several groups reported four independent examples of introns with a splice site that differed from the common intron:
In 1991 by comparing the intron sequences of P120 and CMP genes, IJ Jackson reported the existence of ATATCC (5') and YYCAC (3') splice sites in these introns. The finding indicated a possible novel splicing mechanism.
In 1994, S.L. Hall and R.A Padgett compared the primary sequence of all reports on the four genes mentioned above. The results suggested a new type of introns with ATATCCTT 5’ splice site and YCCAC 3’ splice site and an almost invariant TCCTTAAC near the 3’ end of the introns (so called 3’ upstream element). A search for small nuclear RNA sequences that are complementary to these splice sites, suggested U12 snRNA (matches 3’ sequence) and U11 snRNA (matches 5’sequence) as being putative factors involved in splicing of this new type of introns.
In all these four genes, the pre-mRNA contains other introns whose sequences conform to those of major class introns. Neither the size nor the position of the AT–AC intron within the host gene is conserved.
In 1996, Woan-Yuh Tarn and Joan A. Steitz
described an in vitro system that splices a pre-mRNA substrate containing an AT–AC intron derived from the human P120 gene. Psoralen
cross-linking confirms the base-pairing interaction predicted by Hall and Padgett between the branch site of the pre-mRNA substrate and U12 RNA. Native gel electrophoresis reveals that U11, U12, and U5 snRNPs assemble onto the P120 pre-mRNA to form splicing complexes.
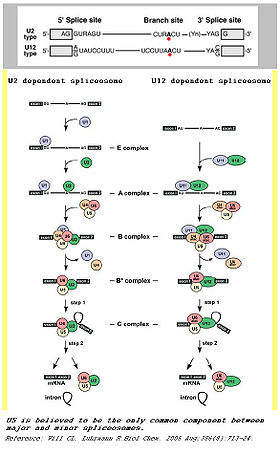
 The main determinants for distinguishing U2- and U12-type introns are 5’ splice site and branch site sequences.
The main determinants for distinguishing U2- and U12-type introns are 5’ splice site and branch site sequences.
Minor spliceosome consists of U11
, U12
, U4atac
, and U6atac
, together with U5
and an unknown number of non-snRNP proteins. The U11, U12 and U4atac/U6atac snRNPs are functional analogs of the U1
, U2
and U4
/U6
snRNPs in major spliceosome. Although the minor U4atac and U6atac snRNAs are functional analogs of U4 and U6, respectively, they share only limited sequence homology (ca. 40%). Furthermore, the sequence of U11 in comparison with U1, as well as U12 compared with U2, are completely unrelated. Despite this fact, the minor U11, U12, U4atac and U6atac snRNAs can be folded into structures similar to U1, U2, U4 and U6, respectively.
Ribonucleoprotein
Ribonucleoprotein is a nucleoprotein that contains RNA, i.e. it is an association that combines ribonucleic acid and protein together. A few known examples include the ribosome, the enzyme telomerase, vault ribonucleoproteins, and small nuclear RNPs , which are implicated in pre-mRNA splicing and...
complex that catalyses the removal (splicing
Splicing (genetics)
In molecular biology and genetics, splicing is a modification of an RNA after transcription, in which introns are removed and exons are joined. This is needed for the typical eukaryotic messenger RNA before it can be used to produce a correct protein through translation...
) of an atypical class of spliceosomal introns (U12-type) from eukaryotic messenger RNA
Messenger RNA
Messenger RNA is a molecule of RNA encoding a chemical "blueprint" for a protein product. mRNA is transcribed from a DNA template, and carries coding information to the sites of protein synthesis: the ribosomes. Here, the nucleic acid polymer is translated into a polymer of amino acids: a protein...
s in plant, insects, vertebrates and some fungi (Rhizopus oryzae
Rhizopus oryzae
Rhizopus oryzae is a fungus that lives worldwide in dead organic matter. An opportunistic human pathogen, it is one causative agent of zygomycosis . The RA 99-880 strain, which was isolated from a fatal infection, had its genome sequenced by the Broad Institute in 2004–2005. R...
). This process is called noncanonical splicing, as opposed to U2-dependent canonical splicing. U12-type introns represent less than 1% of all introns in human cells. However they are found in genes performing essential cellular functions.

Early evidences

Since 1989 till 1991, several groups reported four independent examples of introns with a splice site that differed from the common intron:
- Cartilage matrix protein (CMP/MATN1) gene in human and chicken
- Proliferating cell nucleolar protein P120 (NOL1) gene in human
- mouse Rep3 gene, presumably involved in DNA repair
- Drosophila prospero gene that encodes for a homeobox protein
In 1991 by comparing the intron sequences of P120 and CMP genes, IJ Jackson reported the existence of ATATCC (5') and YYCAC (3') splice sites in these introns. The finding indicated a possible novel splicing mechanism.
In 1994, S.L. Hall and R.A Padgett compared the primary sequence of all reports on the four genes mentioned above. The results suggested a new type of introns with ATATCCTT 5’ splice site and YCCAC 3’ splice site and an almost invariant TCCTTAAC near the 3’ end of the introns (so called 3’ upstream element). A search for small nuclear RNA sequences that are complementary to these splice sites, suggested U12 snRNA (matches 3’ sequence) and U11 snRNA (matches 5’sequence) as being putative factors involved in splicing of this new type of introns.
In all these four genes, the pre-mRNA contains other introns whose sequences conform to those of major class introns. Neither the size nor the position of the AT–AC intron within the host gene is conserved.
In 1996, Woan-Yuh Tarn and Joan A. Steitz
Joan A. Steitz
Joan Argetsinger Steitz is a molecular biologist at Yale University, famed for her discoveries involving RNA, including ground-breaking insights such as that ribosomes interact with mRNA by complementary base pairing and that introns are spliced by snRNPs, small nuclear ribonucleoproteins which...
described an in vitro system that splices a pre-mRNA substrate containing an AT–AC intron derived from the human P120 gene. Psoralen
Psoralen
Psoralen is the parent compound in a family of natural products known as furocoumarins. It is structurally related to coumarin by the addition of a fused furan ring, and may be considered as a derivative of umbelliferone...
cross-linking confirms the base-pairing interaction predicted by Hall and Padgett between the branch site of the pre-mRNA substrate and U12 RNA. Native gel electrophoresis reveals that U11, U12, and U5 snRNPs assemble onto the P120 pre-mRNA to form splicing complexes.
Structure of U12-type introns and minor spliceosome
Although originally referred to as ATAC introns, U2-type introns have GT-AG 5’ and 3’ splice sites while U12-type introns have AT-AC at their 5’ and 3’ ends.
Minor spliceosome consists of U11
U11 spliceosomal RNA
The U11 spliceosomal RNA is a non-coding RNA that together with U4atac/U6atac, U5, and U12 snRNAs and associated proteins, forms a spliceosome that cleaves a divergent class of low-abundance pre-mRNA introns.-References:* [1] * [2]...
, U12
U12 minor spliceosomal RNA
U12 minor spliceosomal RNA is formed from U12 small nuclear , together with U4atac/U6atac, U5, and U11 snRNAs and associated proteins, forms a spliceosome that cleaves a divergent class of low-abundance pre-mRNA introns. Although the U12 sequence is very divergent from that of U2, the two are...
, U4atac
U4atac minor spliceosomal RNA
U4atac minor spliceosomal RNA is a ncRNA which is an essential component of the minor U12-type spliceosome complex. The U12-type spliceosome is required for removal of the rarer class of eukaryotic introns ....
, and U6atac
U6atac minor spliceosomal RNA
U6atac minor spliceosomal RNA is a non-coding RNA which is an essential component of the minor U12-type spliceosome complex. The U12-type spliceosome is required for removal of the rarer class of eukaryotic introns ....
, together with U5
U5 spliceosomal RNA
U5 RNA is a non-coding RNA that is a component of both types of known spliceosome. The precise function of this molecule is unknown, though it is known that the 5' loop is required for splice site selection and p220 binding, and that both the 3' stem-loop and the Sm site are important for Sm...
and an unknown number of non-snRNP proteins. The U11, U12 and U4atac/U6atac snRNPs are functional analogs of the U1
U1 spliceosomal RNA
U1 spliceosomal RNA is the small nuclear RNA component of U1 snRNP , an RNA-protein complex that combines with other snRNPs, unmodified pre-mRNA, and various other proteins to assemble a spliceosome, a large RNA-protein molecular complex upon which splicing of pre-mRNA occurs...
, U2
U2 spliceosomal RNA
U2 spliceosomal RNA is a small nuclear RNA component of the spliceosome . Complementary binding between U2 snRNA and the branchpoint sequence of the intron results in the bulging out of an unpaired adenosine, on the BPS, which initiates a nucleophilic attack at the intronic 5' splice...
and U4
U4 spliceosomal RNA
The U4 small nuclear Ribo-Nucleic Acid is a non-coding RNA component of the major or U2-dependent spliceosome – a eukaryotic molecular machine involved in the splicing of pre-messenger RNA...
/U6
U6 spliceosomal RNA
U6 snRNA is the non-coding small nuclear RNA component of U6 snRNP , an RNA-protein complex that combines with other snRNPs, unmodified pre-mRNA, and various other proteins to assemble a spliceosome, a large RNA-protein molecular complex upon which splicing of pre-mRNA occurs...
snRNPs in major spliceosome. Although the minor U4atac and U6atac snRNAs are functional analogs of U4 and U6, respectively, they share only limited sequence homology (ca. 40%). Furthermore, the sequence of U11 in comparison with U1, as well as U12 compared with U2, are completely unrelated. Despite this fact, the minor U11, U12, U4atac and U6atac snRNAs can be folded into structures similar to U1, U2, U4 and U6, respectively.

