
Polyadenylation
Encyclopedia

RNA
Ribonucleic acid , or RNA, is one of the three major macromolecules that are essential for all known forms of life....
molecule. The poly(A) tail consists of multiple adenosine monophosphate
Adenosine monophosphate
Adenosine monophosphate , also known as 5'-adenylic acid, is a nucleotide that is used as a monomer in RNA. It is an ester of phosphoric acid and the nucleoside adenosine. AMP consists of a phosphate group, the sugar ribose, and the nucleobase adenine...
s; in other words, it is a stretch of RNA that has only adenine
Adenine
Adenine is a nucleobase with a variety of roles in biochemistry including cellular respiration, in the form of both the energy-rich adenosine triphosphate and the cofactors nicotinamide adenine dinucleotide and flavin adenine dinucleotide , and protein synthesis, as a chemical component of DNA...
bases. In eukaryote
Eukaryote
A eukaryote is an organism whose cells contain complex structures enclosed within membranes. Eukaryotes may more formally be referred to as the taxon Eukarya or Eukaryota. The defining membrane-bound structure that sets eukaryotic cells apart from prokaryotic cells is the nucleus, or nuclear...
s, polyadenylation is part of the process that produces mature messenger RNA
Messenger RNA
Messenger RNA is a molecule of RNA encoding a chemical "blueprint" for a protein product. mRNA is transcribed from a DNA template, and carries coding information to the sites of protein synthesis: the ribosomes. Here, the nucleic acid polymer is translated into a polymer of amino acids: a protein...
(mRNA) for translation. It, therefore, forms part of the larger process of gene expression
Gene expression
Gene expression is the process by which information from a gene is used in the synthesis of a functional gene product. These products are often proteins, but in non-protein coding genes such as ribosomal RNA , transfer RNA or small nuclear RNA genes, the product is a functional RNA...
.
The process of polyadenylation begins as the transcription
Transcription (genetics)
Transcription is the process of creating a complementary RNA copy of a sequence of DNA. Both RNA and DNA are nucleic acids, which use base pairs of nucleotides as a complementary language that can be converted back and forth from DNA to RNA by the action of the correct enzymes...
of a gene
Gene
A gene is a molecular unit of heredity of a living organism. It is a name given to some stretches of DNA and RNA that code for a type of protein or for an RNA chain that has a function in the organism. Living beings depend on genes, as they specify all proteins and functional RNA chains...
finishes. The 3'-most
Directionality (molecular biology)
Directionality, in molecular biology and biochemistry, is the end-to-end chemical orientation of a single strand of nucleic acid. The chemical convention of naming carbon atoms in the nucleotide sugar-ring numerically gives rise to a 5′-end and a 3′-end...
segment of the newly-made RNA is first cleaved off by a set of proteins
Protein complex
A multiprotein complex is a group of two or more associated polypeptide chains. If the different polypeptide chains contain different protein domain, the resulting multiprotein complex can have multiple catalytic functions...
; these proteins then synthesize the poly(A) tail at the RNA's 3' end. In some genes, these proteins may add a poly(A) tail at any one of several possible sites. Therefore, polyadenylation can produce more than one transcript from a single gene, similar to alternative splicing
Alternative splicing
Alternative splicing is a process by which the exons of the RNA produced by transcription of a gene are reconnected in multiple ways during RNA splicing...
.
The poly(A) tail is important for the nuclear export, translation, and stability of mRNA. The tail is shortened over time, and, when it is short enough, the mRNA is enzymatically degraded. However, in a few cell types, mRNAs with short poly(A) tails are stored for later activation by re-polyadenylation in the cytosol. In contrast, when polyadenylation occurs in bacteria, it promotes RNA degradation. This is also sometimes the case for eukaryotic non-coding RNA
Non-coding RNA
A non-coding RNA is a functional RNA molecule that is not translated into a protein. Less-frequently used synonyms are non-protein-coding RNA , non-messenger RNA and functional RNA . The term small RNA is often used for short bacterial ncRNAs...
s. The wide distribution of polyadenylation among living organisms indicates that this process evolved
Evolution
Evolution is any change across successive generations in the heritable characteristics of biological populations. Evolutionary processes give rise to diversity at every level of biological organisation, including species, individual organisms and molecules such as DNA and proteins.Life on Earth...
early in the history of life on Earth
Evolutionary history of life
The evolutionary history of life on Earth traces the processes by which living and fossil organisms have evolved since life on Earth first originated until the present day. Earth formed about 4.5 Ga and life appeared on its surface within one billion years...
.
Primer on RNA
- For further information, see RNARNARibonucleic acid , or RNA, is one of the three major macromolecules that are essential for all known forms of life....
and Messenger RNAMessenger RNAMessenger RNA is a molecule of RNA encoding a chemical "blueprint" for a protein product. mRNA is transcribed from a DNA template, and carries coding information to the sites of protein synthesis: the ribosomes. Here, the nucleic acid polymer is translated into a polymer of amino acids: a protein...
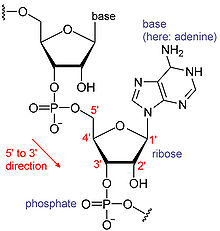
RNAs are a type of large biological molecules, whose individual building blocks are called nucleotides. The name poly(A) tail (for polyadenylic acid tail) reflects the way RNA nucleotides are abbreviated, with a letter for the base the nucleotide contains (A for adenine
Adenine
Adenine is a nucleobase with a variety of roles in biochemistry including cellular respiration, in the form of both the energy-rich adenosine triphosphate and the cofactors nicotinamide adenine dinucleotide and flavin adenine dinucleotide , and protein synthesis, as a chemical component of DNA...
, C for cytosine
Cytosine
Cytosine is one of the four main bases found in DNA and RNA, along with adenine, guanine, and thymine . It is a pyrimidine derivative, with a heterocyclic aromatic ring and two substituents attached . The nucleoside of cytosine is cytidine...
, G for guanine
Guanine
Guanine is one of the four main nucleobases found in the nucleic acids DNA and RNA, the others being adenine, cytosine, and thymine . In DNA, guanine is paired with cytosine. With the formula C5H5N5O, guanine is a derivative of purine, consisting of a fused pyrimidine-imidazole ring system with...
and U for uracil
Uracil
Uracil is one of the four nucleobases in the nucleic acid of RNA that are represented by the letters A, G, C and U. The others are adenine, cytosine, and guanine. In RNA, uracil binds to adenine via two hydrogen bonds. In DNA, the uracil nucleobase is replaced by thymine.Uracil is a common and...
). RNAs are produced (transcribed
Transcription (genetics)
Transcription is the process of creating a complementary RNA copy of a sequence of DNA. Both RNA and DNA are nucleic acids, which use base pairs of nucleotides as a complementary language that can be converted back and forth from DNA to RNA by the action of the correct enzymes...
) from a DNA
DNA
Deoxyribonucleic acid is a nucleic acid that contains the genetic instructions used in the development and functioning of all known living organisms . The DNA segments that carry this genetic information are called genes, but other DNA sequences have structural purposes, or are involved in...
template. By convention, RNA sequences are written in a 5' to 3' direction. The 5' end is the part of the RNA molecule that is transcribed first, and the 3' end is transcribed last. The 3' end is also where the poly(A) tail is found on polyadenylated RNAs.
Messenger RNA (mRNA) is RNA that has a coding region that acts as a template for protein synthesis (translation). The rest of the mRNA, the untranslated regions, tune how active the mRNA is. There are also many RNAs that are not translated, called non-coding RNAs. Like the untranslated regions, many of these non-coding RNAs have regulatory roles.
Function
In nuclear polyadenylation, a poly(A) tail is added to an RNA at the end of transcription. On mRNAs, the poly(A) tail protects the mRNA molecule from enzymatic degradation in the cytoplasmCytoplasm
The cytoplasm is a small gel-like substance residing between the cell membrane holding all the cell's internal sub-structures , except for the nucleus. All the contents of the cells of prokaryote organisms are contained within the cytoplasm...
and aids in transcription termination, export of the mRNA from the nucleus, and translation. Almost all eukaryotic mRNAs are polyadenylated, with the exception of animal replication-dependent histone
Histone
In biology, histones are highly alkaline proteins found in eukaryotic cell nuclei that package and order the DNA into structural units called nucleosomes. They are the chief protein components of chromatin, acting as spools around which DNA winds, and play a role in gene regulation...
mRNAs. These are the only mRNAs in eukaryotes that lack a poly(A) tail, ending instead in a stem-loop
Histone 3' UTR stem-loop
The Histone 3' UTR stem-loop is an RNA element involved in nucleocytoplasmic transport of the histone mRNAs, and in the regulation of stability and of translation efficiency in the cytoplasm...
structure followed by a purine-rich sequence, termed histone downstream element, that directs where the RNA is cut so that the 3' end of the histone mRNA is formed.
Many eukaryotic non-coding RNAs are always polyadenylated at the end of transcription. There are small RNAs where the poly(A) tail is seen only in intermediary forms and not in the mature RNA as the ends are removed during processing, the notable ones being microRNAs. But, for many long noncoding RNA
Long noncoding RNA
Long non-coding RNAs are generally considered as non-protein coding transcripts longer than 200 nucleotides. This limit is due to practical considerations including the separation of RNAs in common experimental protocols...
s – a seemingly large group of regulatory
Regulation of gene expression
Gene modulation redirects here. For information on therapeutic regulation of gene expression, see therapeutic gene modulation.Regulation of gene expression includes the processes that cells and viruses use to regulate the way that the information in genes is turned into gene products...
RNAs that, for example, includes the RNA Xist, which mediates X chromosome inactivation – a poly(A) tail is part of the mature RNA.
Mechanism
| Proteins involved: CPSF Cleavage and polyadenylation specificity factor Cleavage and polyadenylation specificity factor is involved in the cleavage of the 3' signaling region from a newly synthesized pre-messenger RNA molecule in the process of gene transcription... : cleavage/polyadenylation specificity factor CstF Cleavage stimulatory factor Cleavage stimulatory factor or cleavage stimulation factor is a heterotrimeric protein of about 200 kilodaltons that is involved in the cleavage of the 3' signaling region from a newly synthesized pre-messenger RNA molecule... : cleavage stimulation factor PAP Polynucleotide adenylyltransferase In enzymology, a polynucleotide adenylyltransferase is an enzyme that catalyzes the chemical reactionThus, the two substrates of this enzyme are ATP and RNA, whereas its two products are diphosphate and RNA with an extra adenosine nucleotide at its 3' end.... : polyadenylate polymerase PAB2 PABPN1 Polyadenylate-binding protein 2 also known as polyadenylate-binding nuclear protein 1 is a protein that in humans is encoded by the PABPN1 gene.- Function :... : polyadenylate binding protein 2 CFI Cleavage factor Cleavage factors are two closely related proteins involved in the cleavage of the 3' signaling region from a newly synthesized pre-messenger RNA molecule in the process of gene transcription. The cleavage is the first step in adding a polyadenine tail to the pre-mRNA, which is one of the necessary... : cleavage factor I CFII Cleavage factor Cleavage factors are two closely related proteins involved in the cleavage of the 3' signaling region from a newly synthesized pre-messenger RNA molecule in the process of gene transcription. The cleavage is the first step in adding a polyadenine tail to the pre-mRNA, which is one of the necessary... : cleavage factor II |
The polyadenylation machinery in the nucleus of eukaryotes works on products of RNA polymerase II
RNA polymerase II
RNA polymerase II is an enzyme found in eukaryotic cells. It catalyzes the transcription of DNA to synthesize precursors of mRNA and most snRNA and microRNA. A 550 kDa complex of 12 subunits, RNAP II is the most studied type of RNA polymerase...
, such as precursor mRNA
Precursor mRNA
Precursor mRNA is an immature single strand of messenger ribonucleic acid . pre-mRNA is synthesized from a DNA template in the cell nucleus by transcription. Pre-mRNA comprises the bulk of heterogeneous nuclear RNA...
. Here, a multi-protein complex (see components on the right) cleaves the 3'-most part of a newly produced RNA and polyadenylates the end produced by this cleavage. The cleavage is catalysed by the enzyme CPSF and occurs 10–30 nucleotides downstream of its binding site. This site is often the sequence AAUAAA on the RNA, but variants of it that bind more weakly to CPSF exist. Two other proteins add specificity to the binding to an RNA: CstF and CFI. CstF binds to a GU-rich region further downstream of CPSF's site. CFI recognises a third site on the RNA (a set of UGUAA sequences in mammals) and can recruit CPSF even if the AAUAAA sequence is missing. The polyadenylation signal – the sequence motif recognised by the RNA cleavage complex – varies between groups of eukaryotes. Most human polyadenylation sites contain the AAUAAA sequence, but this sequence is less common in plants and fungi.
The RNA is cleaved right after transcription, as CstF also binds to RNA polymerase II. Cleavage also involves the protein CFII, though it is unknown how. The cleavage site associated with a polyadenylation signal can vary up to some 50 nucleotides.
When the RNA is cleaved, polyadenylation starts, catalysed by polyadenylate polymerase. Polyadenylate polymerase builds the poly(A) tail by adding adenosine monophosphate
Adenosine monophosphate
Adenosine monophosphate , also known as 5'-adenylic acid, is a nucleotide that is used as a monomer in RNA. It is an ester of phosphoric acid and the nucleoside adenosine. AMP consists of a phosphate group, the sugar ribose, and the nucleobase adenine...
units from adenosine triphosphate
Adenosine triphosphate
Adenosine-5'-triphosphate is a multifunctional nucleoside triphosphate used in cells as a coenzyme. It is often called the "molecular unit of currency" of intracellular energy transfer. ATP transports chemical energy within cells for metabolism...
to the RNA, cleaving off pyrophosphate
Pyrophosphate
In chemistry, the anion, the salts, and the esters of pyrophosphoric acid are called pyrophosphates. Any salt or ester containing two phosphate groups is called a diphosphate. As a food additive, diphosphates are known as E450.- Chemistry :...
. Another protein, PAB2, binds to the new, short poly(A) tail and increases the affinity of polyadenylate polymerase for the RNA. When the poly(A) tail is approximately 250 nucleotide
Nucleotide
Nucleotides are molecules that, when joined together, make up the structural units of RNA and DNA. In addition, nucleotides participate in cellular signaling , and are incorporated into important cofactors of enzymatic reactions...
s long the enzyme can no longer bind to CPSF and polyadenylation stops, thus determining the length of the poly(A) tail. CPSF is in contact with RNA polymerase II, allowing it to tell the polymerase to terminate transcription. The polyadenylation machinery is also physically linked to the spliceosome
Spliceosome
A spliceosome is a complex of snRNA and protein subunits that removes introns from a transcribed pre-mRNA segment. This process is generally referred to as splicing.-Composition:...
, a complex that removes intron
Intron
An intron is any nucleotide sequence within a gene that is removed by RNA splicing to generate the final mature RNA product of a gene. The term intron refers to both the DNA sequence within a gene, and the corresponding sequence in RNA transcripts. Sequences that are joined together in the final...
s from RNAs.
Downstream effects
The poly(A) tail acts as the binding site for poly(A)-binding proteinPoly(A)-binding protein
Poly-binding protein is a RNA-binding protein which binds to the poly tail of mRNA. The poly tail is located on the 3' end of mRNA...
. Poly(A)-binding protein promotes export from the nucleus and translation, and inhibits degradation. This protein binds to the poly(A) tail prior to mRNA export from the nucleus and in yeast also recruits poly(A) nuclease, an enzyme that shortens the poly(A) tail and allows the export of the mRNA. Poly(A)-binding protein is exported to the cytoplasm with the RNA. mRNAs that are not exported are degraded by the exosome
Exosome complex
The exosome complex is a multi-protein complex capable of degrading various types of RNA molecules...
. Poly(A)-binding protein also can bind to, and thus recruit, several proteins that affect translation, one of these is initiation factor
Eukaryotic initiation factor
Eukaryotic initiation factors are proteins involved in the initiation phase of eukaryotic translation. They function in forming a complex with the 40S ribosomal subunit and Met-tRNAi called the 43S preinitation complex , recognizing the 5' cap structure of mRNA and recruiting the 43S PIC to mRNA,...
-4G, which in turn recruits the 40S
40S
40S is the small subunit of eukaryotic ribosomes.It interacts with the internal ribosome entry site of the hepatitis C virus.The following is a list of proteins contained in the 40S ribosome:...
ribosomal subunit. However, a poly(A) tail is not required for the translation of all mRNAs.
Deadenylation
In eukaryotic somatic cellSomatic cell
A somatic cell is any biological cell forming the body of an organism; that is, in a multicellular organism, any cell other than a gamete, germ cell, gametocyte or undifferentiated stem cell...
s, the poly(A) tail of most mRNAs in the cytoplasm gradually get shorter, and mRNAs with shorter poly(A) tail are translated less and degraded sooner. However, it can take many hours before an mRNA is degraded. This deadenylation and degradation process can be accelerated by microRNAs complementary to the 3' untranslated region of an mRNA. In immature egg cells
Oocyte
An oocyte, ovocyte, or rarely ocyte, is a female gametocyte or germ cell involved in reproduction. In other words, it is an immature ovum, or egg cell. An oocyte is produced in the ovary during female gametogenesis. The female germ cells produce a primordial germ cell which undergoes a mitotic...
, mRNAs with shortened poly(A) tails are not degraded, but are instead stored without being translated. They are then activated by cytoplasmic polyadenylation after fertilisation, during egg activation.
In animals, poly(A) ribonuclease (PARN
PARN
Poly-specific ribonuclease PARN also known as polyadenylate-specific ribonuclease or as deadenylating nuclease is an enzyme that in humans is encoded by the PARN gene.- Function :...
) can bind to the 5' cap
5' cap
The 5' cap is a specially altered nucleotide on the 5' end of precursor messenger RNA and some other primary RNA transcripts as found in eukaryotes. The process of 5' capping is vital to creating mature messenger RNA, which is then able to undergo translation...
and remove nucleotides from the poly(A) tail. The level of access to the 5' cap and poly(A) tail is important in controlling how soon the mRNA is degraded. PARN deadenylates less if the RNA is bound by the initiation factors 4E (at the 5' cap) and 4G (at the poly(A) tail), which is why translation reduces deadenylation. The rate of deadenylation may also be regulated by RNA-binding proteins. Once the poly(A) tail is removed, the decapping complex removes the 5' cap, leading to a degradation of the RNA. Several other enzymes that seem to be involved in deadenylation have been identified in yeast.
Alternative polyadenylation
Many protein-coding genes have more than one polyadenylation site, so a gene can code for several mRNAs that differ in their 3' end. Since alternative polyadenylation changes the length of the 3' untranslated regionThree prime untranslated region
In molecular genetics, the three prime untranslated region is a particular section of messenger RNA . It is preceeded by the coding region....
, it can change which binding sites for microRNAs the 3' untranslated region contains. MicroRNAs tend to repress translation and promote degradation of the mRNAs they bind to, although there are examples of microRNAs that stabilise transcripts. Alternative polyadenylation can also shorten the coding region, thus making the mRNA code for a different protein, but this is much less common than just shortening the 3' untranslated region.
The choice of poly(A) site can be influenced by extracellular stimuli and depends on the expression of the proteins that take part in polyadenylation. For example, the expression of CstF-64
CSTF2
Cleavage stimulation factor 64 kDa subunit is a protein that in humans is encoded by the CSTF2 gene.-Interactions:CSTF2 has been shown to interact with CSTF3, SUB1, SYMPK, BARD1 and BRCA1.-Further reading:...
, a subunit of cleavage stimulatory factor
Cleavage stimulatory factor
Cleavage stimulatory factor or cleavage stimulation factor is a heterotrimeric protein of about 200 kilodaltons that is involved in the cleavage of the 3' signaling region from a newly synthesized pre-messenger RNA molecule...
(CstF), increases in macrophage
Macrophage
Macrophages are cells produced by the differentiation of monocytes in tissues. Human macrophages are about in diameter. Monocytes and macrophages are phagocytes. Macrophages function in both non-specific defense as well as help initiate specific defense mechanisms of vertebrate animals...
s in response to lipopolysaccharide
Lipopolysaccharide
Lipopolysaccharides , also known as lipoglycans, are large molecules consisting of a lipid and a polysaccharide joined by a covalent bond; they are found in the outer membrane of Gram-negative bacteria, act as endotoxins and elicit strong immune responses in animals.-Functions:LPS is the major...
s (a group of bacterial compounds that trigger an immune response). This results in the selection of weak poly(A) sites and thus shorter transcripts. This removes regulatory elements in the 3' untranslated regions of mRNAs for defense-related products like lysozyme
Lysozyme
Lysozyme, also known as muramidase or N-acetylmuramide glycanhydrolase, are glycoside hydrolases, enzymes that damage bacterial cell walls by catalyzing hydrolysis of 1,4-beta-linkages between N-acetylmuramic acid and N-acetyl-D-glucosamine residues in a peptidoglycan and between...
and TNF-α. These mRNAs then have longer half-lives and produce more of these proteins. RNA-binding proteins other than those in the polyadenylation machinery can also affect whether a polyadenyation site is used,as can DNA methylation
DNA methylation
DNA methylation is a biochemical process that is important for normal development in higher organisms. It involves the addition of a methyl group to the 5 position of the cytosine pyrimidine ring or the number 6 nitrogen of the adenine purine ring...
near the polyadenylation signal.
Cytoplasmic polyadenylation
There is polyadenylation in the cytosol of some animal cell types, namely in the germ line, during early embryogenesisEmbryogenesis
Embryogenesis is the process by which the embryo is formed and develops, until it develops into a fetus.Embryogenesis starts with the fertilization of the ovum by sperm. The fertilized ovum is referred to as a zygote...
and in post-synaptic
Chemical synapse
Chemical synapses are specialized junctions through which neurons signal to each other and to non-neuronal cells such as those in muscles or glands. Chemical synapses allow neurons to form circuits within the central nervous system. They are crucial to the biological computations that underlie...
sites of nerve cells
Neuron
A neuron is an electrically excitable cell that processes and transmits information by electrical and chemical signaling. Chemical signaling occurs via synapses, specialized connections with other cells. Neurons connect to each other to form networks. Neurons are the core components of the nervous...
. This lengthens the poly(A) tail of an mRNA with a shortened poly(A) tail, so that the mRNA will be translated. These shortened poly(A) tails are often less than 20 nucleotides, and are lengthened to around 80–150 nucleotides.
In the early mouse embryo, cytoplasmic polyadenylation of maternal RNAs from the egg cell allows the cell to survive and grow even though transcription does not start until the middle of the 2-cell stage (4-cell stage in human). In the brain, cytoplasmic polyadenylation is active during learning and could play a role in long-term potentiation
Long-term potentiation
In neuroscience, long-term potentiation is a long-lasting enhancement in signal transmission between two neurons that results from stimulating them synchronously. It is one of several phenomena underlying synaptic plasticity, the ability of chemical synapses to change their strength...
, which is the strengthening of the signal transmission from a nerve cell to another in response to nerve impulses and is important for learning and memory formation.
Cytoplasmic polyadenylation requires the RNA-binding proteins CPSF
Cleavage and polyadenylation specificity factor
Cleavage and polyadenylation specificity factor is involved in the cleavage of the 3' signaling region from a newly synthesized pre-messenger RNA molecule in the process of gene transcription...
and CPEB
CPEB
CPEB, or cytoplasmic polyadenylation element binding protein, is a highly conserved RNA-binding protein that promotes the elongation of the polyadenine tail of messenger RNA. CPEB most commonly activates the target RNA for translation, but can also act as a repressor, dependent on its...
, and can involve other RNA-binding proteins like Pumilio
PUM1
Pumilio homolog 1 is a protein that in humans is encoded by the PUM1 gene.- Function :This gene encodes a member of the PUF family, evolutionarily conserved RNA-binding proteins related to the Pumilio proteins of Drosophila and the fem-3 mRNA binding factor proteins of C. elegans...
. Depending on the cell type, the polymerase can be the same type of polyadenylate polymerase (PAP) that is used in the nuclear process, or the cytoplasmic polymerase GLD-2.
Tagging for degradation in eukaryotes
For many non-coding RNANon-coding RNA
A non-coding RNA is a functional RNA molecule that is not translated into a protein. Less-frequently used synonyms are non-protein-coding RNA , non-messenger RNA and functional RNA . The term small RNA is often used for short bacterial ncRNAs...
s, including tRNA
Transfer RNA
Transfer RNA is an adaptor molecule composed of RNA, typically 73 to 93 nucleotides in length, that is used in biology to bridge the three-letter genetic code in messenger RNA with the twenty-letter code of amino acids in proteins. The role of tRNA as an adaptor is best understood by...
, rRNA
Ribosomal RNA
Ribosomal ribonucleic acid is the RNA component of the ribosome, the enzyme that is the site of protein synthesis in all living cells. Ribosomal RNA provides a mechanism for decoding mRNA into amino acids and interacts with tRNAs during translation by providing peptidyl transferase activity...
, snRNA
Small nuclear RNA
Small nuclear ribonucleic acid is a class of small RNA molecules that are found within the nucleus of eukaryotic cells. They are transcribed by RNA polymerase II or RNA polymerase III and are involved in a variety of important processes such as RNA splicing , regulation of transcription factors ...
, and snoRNA
SnoRNA
Small nucleolar RNAs are a class of small RNA molecules that primarily guide chemical modifications of other RNAs, mainly ribosomal RNAs, transfer RNAs and small nuclear RNAs...
, polyadenylation is a way of marking the RNA for degradation, in at least yeast
Saccharomyces cerevisiae
Saccharomyces cerevisiae is a species of yeast. It is perhaps the most useful yeast, having been instrumental to baking and brewing since ancient times. It is believed that it was originally isolated from the skin of grapes...
. This polyadenylation is done in the nucleus by the TRAMP complex
TRAMP complex
The TRAMP complex is a multi-protein complex consisting of the RNA helicase Mtr4, a poly polymerase and a zinc knuckle protein...
, which adds a tail that is around 40 nucleotides long to the 3' end. The RNA is then degraded by the exosome
Exosome complex
The exosome complex is a multi-protein complex capable of degrading various types of RNA molecules...
. Poly(A) tails have also been found on human rRNA fragments, both the form of homopolymeric (A only) and heterpolymeric (mostly A) tails.
In prokaryotes and organelles
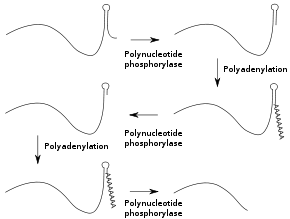
Degradosome
The degradosome is a multi-protein complex present in most bacteria that is involved in the processing of ribosomal RNA and the degradation of messenger RNA. It contains the proteins RNA helicase B, RNase E and Polynucleotide phosphorylase....
, which contains two RNA-degrading enzymes: polynucleotide phosphorylase and RNase E. Polynucleotide phosphorylase binds to the 3' end of RNAs and the 3' extension provided by the poly(A) tail allows it to bind to the RNAs whose secondary structure
Secondary structure
In biochemistry and structural biology, secondary structure is the general three-dimensional form of local segments of biopolymers such as proteins and nucleic acids...
would otherwise block the 3' end. Successive rounds of polyadenylation and degradation of the 3' end by polynucleotide phosphorylase allows the degradosome to overcome these secondary structures. The poly(A) tail can also recruit RNases that cut the RNA in two. These bacterial poly(A) tails are about 30 nucleotides long.
In as different groups as animals and trypanosomes, the mitochondria
Mitochondrion
In cell biology, a mitochondrion is a membrane-enclosed organelle found in most eukaryotic cells. These organelles range from 0.5 to 1.0 micrometers in diameter...
contain both stabilising and destabilising poly(A) tails. Destabilising polyadenylation targets both mRNA and noncoding RNAs. The poly(A) tails are 43 nucleotides long on average. The stabilising ones start at the stop codon, and without them the stop codon (UAA) is not complete as the genome only encodes the U or UA part. Plant mitochondria have only destabilising polyadenylation, and yeast mitochondria have no polyadenylation at all.
While many bacteria and mitochondria have polyadenylate polymerases, they also have another type of polyadenylation, performed by polynucleotide phosphorylase
Polynucleotide phosphorylase
Polynucleotide Phosphorylase is a bifunctional enzyme with a phosphorolytic 3' to 5' exoribonuclease activity and a 3'-terminal oligonucleotide polymerase activity. It is involved on mRNA processing and degradation in bacteria, plants, and in humans.In humans, the enzyme is encoded by the gene...
itself. This enzyme is found in bacteria, mitochondria, plastid
Plastid
Plastids are major organelles found in the cells of plants and algae. Plastids are the site of manufacture and storage of important chemical compounds used by the cell...
s and as a constituent of the archeal exosome (in those archaea
Archaea
The Archaea are a group of single-celled microorganisms. A single individual or species from this domain is called an archaeon...
that have an exosome
Exosome complex
The exosome complex is a multi-protein complex capable of degrading various types of RNA molecules...
). It can synthesise a 3' extension where the vast majority of the bases are adenines. Like in bacteria, polyadenylation by polynucleotide phosphorylase promotes degradation of the RNA in plastids and likely also archaea.
Evolution
Although polyadenylation is seen in almost all organisms, it is not universal. However, the wide distribution of this modification and the fact that it is present in organisms from all three domainsDomain (biology)
In biological taxonomy, a domain is the highest taxonomic rank of organisms, higher than a kingdom. According to the three-domain system of Carl Woese, introduced in 1990, the Tree of Life consists of three domains: Archaea, Bacteria and Eukarya...
of life implies that the last universal common ancestor
Last universal ancestor
The last universal ancestor , also called the last universal common ancestor , or the cenancestor, is the most recent organism from which all organisms now living on Earth descend. Thus it is the most recent common ancestor of all current life on Earth...
of all living organisms, it is presumed, had some form of polyadenylation system. A few organisms do not polyadenylate mRNA, which implies that they have lost their polyadenylation machineries during evolution. Although no examples of eukaryotes that lack polyadenylation are known, mRNAs from the bacterium Mycoplasma gallisepticum
Mycoplasma gallisepticum
Mycoplasma gallisepticum is a bacterium belonging to the class Mollicutes and the family Mycoplasmataceae. It is the causative agent of chronic respiratory disease in chickens and infectious sinusitis in turkeys, chickens, game birds, pigeons, and passerine birds of all ages.-Transmission:MG is...
and the salt-tolerant archaean Haloferax volcanii
Haloferax
In taxonomy, Haloferax is a genus of the Halobacteriaceae.- Description and Significance :Haloferax volcanii is a species of halophile which exists in extreme saline environments. Recently an isolate of this species was studied by researchers at University of California-Berkeley as part of a...
lack this modification.
The most ancient polyadenylating enzyme is polynucleotide phosphorylase
Polynucleotide phosphorylase
Polynucleotide Phosphorylase is a bifunctional enzyme with a phosphorolytic 3' to 5' exoribonuclease activity and a 3'-terminal oligonucleotide polymerase activity. It is involved on mRNA processing and degradation in bacteria, plants, and in humans.In humans, the enzyme is encoded by the gene...
. This enzyme is part of both the bacterial degradosome
Degradosome
The degradosome is a multi-protein complex present in most bacteria that is involved in the processing of ribosomal RNA and the degradation of messenger RNA. It contains the proteins RNA helicase B, RNase E and Polynucleotide phosphorylase....
and the archaeal exosome
Exosome complex
The exosome complex is a multi-protein complex capable of degrading various types of RNA molecules...
, two closely related complexes that recycle RNA into nucleotides. This enzyme degrades RNA by attacking the bond between the 3'-most nucleotides with a phosphate, breaking off a diphosphate nucleotide. This reaction is reversible, and so the enzyme can also extend RNA with more nucleotides. The heteropolymeric tail added by polynucleotide phosphorylase is very rich in adenine. The choice of adenine is most likely the result of higher ADP
Adenosine diphosphate
Adenosine diphosphate, abbreviated ADP, is a nucleoside diphosphate. It is an ester of pyrophosphoric acid with the nucleoside adenosine. ADP consists of the pyrophosphate group, the pentose sugar ribose, and the nucleobase adenine....
concentrations than other nucleotides as a result of using ATP
Adenosine triphosphate
Adenosine-5'-triphosphate is a multifunctional nucleoside triphosphate used in cells as a coenzyme. It is often called the "molecular unit of currency" of intracellular energy transfer. ATP transports chemical energy within cells for metabolism...
as an energy currency, making it more likely to be incorporated in this tail in early lifeforms. It has been suggested that the involvement of adenine-rich tails in RNA degradation prompted the later evolution of polyadenylate polymerases (the enzymes that produce poly(A) tails with no other nucleotides in them).
Polyadenylate polymerases are not as ancient. They have separately evolved in both bacteria and eukaryotes from CCA-adding enzyme
TRNT1
tRNA-nucleotidyltransferase 1, mitochondrial is an enzyme that in humans is encoded by the TRNT1 gene.This enzyme adds the nucleotide sequence CCA to the 3' end of tRNA, using ATP and CTP as substrates. The sequence creates the binding site for an amino acid.-Further reading:...
, which is the enzyme that completes the 3' ends of tRNAs
Transfer RNA
Transfer RNA is an adaptor molecule composed of RNA, typically 73 to 93 nucleotides in length, that is used in biology to bridge the three-letter genetic code in messenger RNA with the twenty-letter code of amino acids in proteins. The role of tRNA as an adaptor is best understood by...
. Its catalytic domain is homologous to that of other polymerase
Polymerase
A polymerase is an enzyme whose central function is associated with polymers of nucleic acids such as RNA and DNA.The primary function of a polymerase is the polymerization of new DNA or RNA against an existing DNA or RNA template in the processes of replication and transcription...
s. It is presumed that the horizontal transfer of bacterial CCA-adding enzyme to eukaryotes allowed the archaeal-like CCA-adding enzyme to switch function to a poly(A) polymerase. Some lineages, like archaea
Archaea
The Archaea are a group of single-celled microorganisms. A single individual or species from this domain is called an archaeon...
and cyanobacteria, never evolved a polyadenylate polymerase.
History
Polyadenylation was first identified in 1960 as an enzymatic activityEnzyme
Enzymes are proteins that catalyze chemical reactions. In enzymatic reactions, the molecules at the beginning of the process, called substrates, are converted into different molecules, called products. Almost all chemical reactions in a biological cell need enzymes in order to occur at rates...
in extracts made from cell nuclei that could polymerise ATP, but not ADP, into polyadenine. Although identified in many types of cells, this activity had no known function until 1971, when poly(A) sequences were found in mRNAs. The only function of these sequences was thought at first to be protection of the 3' end of the RNA from nucleases, but later the specific roles of polyadenylation in nuclear export and translation were identified. The polymerases responsible for polyadenylation were first purified and characterized in the 1960s and 1970s, but the large number of accessory proteins that control this process were discovered only in the early 1990s.

