.gif)
Force field (chemistry)
Encyclopedia
Parameter
Parameter from Ancient Greek παρά also “para” meaning “beside, subsidiary” and μέτρον also “metron” meaning “measure”, can be interpreted in mathematics, logic, linguistics, environmental science and other disciplines....
s of mathematical functions
Function (mathematics)
In mathematics, a function associates one quantity, the argument of the function, also known as the input, with another quantity, the value of the function, also known as the output. A function assigns exactly one output to each input. The argument and the value may be real numbers, but they can...
used to describe the potential energy
Potential energy
In physics, potential energy is the energy stored in a body or in a system due to its position in a force field or due to its configuration. The SI unit of measure for energy and work is the Joule...
of a system of particles (typically molecule
Molecule
A molecule is an electrically neutral group of at least two atoms held together by covalent chemical bonds. Molecules are distinguished from ions by their electrical charge...
s and atom
Atom
The atom is a basic unit of matter that consists of a dense central nucleus surrounded by a cloud of negatively charged electrons. The atomic nucleus contains a mix of positively charged protons and electrically neutral neutrons...
s). Force field functions and parameter sets are derived from both experimental work and high-level quantum mechanical
Quantum mechanics
Quantum mechanics, also known as quantum physics or quantum theory, is a branch of physics providing a mathematical description of much of the dual particle-like and wave-like behavior and interactions of energy and matter. It departs from classical mechanics primarily at the atomic and subatomic...
calculations. "All-atom" force fields provide parameters for every type of atom in a system, including hydrogen
Hydrogen
Hydrogen is the chemical element with atomic number 1. It is represented by the symbol H. With an average atomic weight of , hydrogen is the lightest and most abundant chemical element, constituting roughly 75% of the Universe's chemical elemental mass. Stars in the main sequence are mainly...
, while "united-atom" force fields treat the hydrogen and carbon
Carbon
Carbon is the chemical element with symbol C and atomic number 6. As a member of group 14 on the periodic table, it is nonmetallic and tetravalent—making four electrons available to form covalent chemical bonds...
atoms in methyl and methylene
Methylene
Methylene is a chemical species in which a carbon atom is bonded to two hydrogen atoms. Three different possibilities present themselves:* the -CH2- substituent group: e.g., dichloromethane ....
groups as a single interaction center. "Coarse-grained" force fields, which are frequently used in long-time simulations of protein
Protein
Proteins are biochemical compounds consisting of one or more polypeptides typically folded into a globular or fibrous form, facilitating a biological function. A polypeptide is a single linear polymer chain of amino acids bonded together by peptide bonds between the carboxyl and amino groups of...
s, provide even more crude representations for increased computational efficiency.
The usage of the term "force field" in chemistry and computational biology differs from the standard usage in physics
Physics
Physics is a natural science that involves the study of matter and its motion through spacetime, along with related concepts such as energy and force. More broadly, it is the general analysis of nature, conducted in order to understand how the universe behaves.Physics is one of the oldest academic...
. In chemistry it is a system of potential energy functions rather than the gradient
Gradient
In vector calculus, the gradient of a scalar field is a vector field that points in the direction of the greatest rate of increase of the scalar field, and whose magnitude is the greatest rate of change....
of a scalar potential
Scalar potential
A scalar potential is a fundamental concept in vector analysis and physics . The scalar potential is an example of a scalar field...
, as defined in physics.
Functional form
Chemical bond
A chemical bond is an attraction between atoms that allows the formation of chemical substances that contain two or more atoms. The bond is caused by the electromagnetic force attraction between opposite charges, either between electrons and nuclei, or as the result of a dipole attraction...
terms relating to atoms that are linked by covalent bond
Covalent bond
A covalent bond is a form of chemical bonding that is characterized by the sharing of pairs of electrons between atoms. The stable balance of attractive and repulsive forces between atoms when they share electrons is known as covalent bonding....
s, and nonbonded (also called "noncovalent") terms describing the long-range electrostatic and van der Waals
Van der Waals force
In physical chemistry, the van der Waals force , named after Dutch scientist Johannes Diderik van der Waals, is the sum of the attractive or repulsive forces between molecules other than those due to covalent bonds or to the electrostatic interaction of ions with one another or with neutral...
forces. The specific decomposition of the terms depends on the force field, but a general form for the total energy in an additive force field can be written as
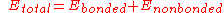
where the components of the covalent and noncovalent contributions are given by the following summations:


The bond and angle terms are usually modeled as harmonic oscillator
Harmonic oscillator
In classical mechanics, a harmonic oscillator is a system that, when displaced from its equilibrium position, experiences a restoring force, F, proportional to the displacement, x: \vec F = -k \vec x \, where k is a positive constant....
s in force fields that do not allow bond breaking. A more realistic description of a covalent bond at higher stretching is provided by the more expensive Morse potential
Morse potential
The Morse potential, named after physicist Philip M. Morse, is a convenient model for the potential energy of a diatomic molecule. It is a better approximation for the vibrational structure of the molecule than the quantum harmonic oscillator because it explicitly includes the effects of bond...
. The functional form for the rest of the bonded terms is highly variable. Proper dihedral potentials are usually included. Additionally, "improper torsional" terms may be added to enforce the planarity of aromatic rings and other conjugated system
Conjugated system
In chemistry, a conjugated system is a system of connected p-orbitals with delocalized electrons in compounds with alternating single and multiple bonds, which in general may lower the overall energy of the molecule and increase stability. Lone pairs, radicals or carbenium ions may be part of the...
s, and "cross-terms" that describe coupling of different internal variables, such as angles and bond lengths. Some force fields also include explicit terms for hydrogen bond
Hydrogen bond
A hydrogen bond is the attractive interaction of a hydrogen atom with an electronegative atom, such as nitrogen, oxygen or fluorine, that comes from another molecule or chemical group. The hydrogen must be covalently bonded to another electronegative atom to create the bond...
s.
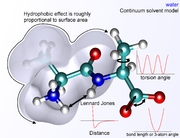
The nonbonded terms are most computationally intensive because they include many more interactions per atom. A popular choice is to limit interactions to pairwise energies. The van der Waals term is usually computed with a Lennard-Jones potential
Lennard-Jones potential
The Lennard-Jones potential is a mathematically simple model that approximates the interaction between a pair of neutral atoms or molecules. A form of the potential was first proposed in 1924 by John Lennard-Jones...
and the electrostatic term with Coulomb's law
Coulomb's law
Coulomb's law or Coulomb's inverse-square law, is a law of physics describing the electrostatic interaction between electrically charged particles. It was first published in 1785 by French physicist Charles Augustin de Coulomb and was essential to the development of the theory of electromagnetism...
, although both can be buffered or scaled by a constant factor to account for electronic polarizability
Polarizability
Polarizability is the measure of the change in a molecule's electron distribution in response to an applied electric field, which can also be induced by electric interactions with solvents or ionic reagents. It is a property of matter...
and produce better agreement with experimental observations.
Parameterization
In addition to the functional form of the potentials, a force field defines a set of parameters for each type of atom. For example, a force field would include distinct parameters for an oxygenOxygen
Oxygen is the element with atomic number 8 and represented by the symbol O. Its name derives from the Greek roots ὀξύς and -γενής , because at the time of naming, it was mistakenly thought that all acids required oxygen in their composition...
atom in a carbonyl
Carbonyl
In organic chemistry, a carbonyl group is a functional group composed of a carbon atom double-bonded to an oxygen atom: C=O. It is common to several classes of organic compounds, as part of many larger functional groups....
functional group
Functional group
In organic chemistry, functional groups are specific groups of atoms within molecules that are responsible for the characteristic chemical reactions of those molecules. The same functional group will undergo the same or similar chemical reaction regardless of the size of the molecule it is a part of...
and in a hydroxyl
Hydroxyl
A hydroxyl is a chemical group containing an oxygen atom covalently bonded with a hydrogen atom. In inorganic chemistry, the hydroxyl group is known as the hydroxide ion, and scientists and reference works generally use these different terms though they refer to the same chemical structure in...
group. The typical parameter set includes values for atomic mass
Atomic mass
The atomic mass is the mass of a specific isotope, most often expressed in unified atomic mass units. The atomic mass is the total mass of protons, neutrons and electrons in a single atom....
, van der Waals radius
Van der Waals radius
The van der Waals radius, r, of an atom is the radius of an imaginary hard sphere which can be used to model the atom for many purposes. It is named after Johannes Diderik van der Waals, winner of the 1910 Nobel Prize in Physics, as he was the first to recognise that atoms had a finite size and to...
, and partial charge
Partial charge
A partial charge is a charge with an absolute value of less than one elementary charge unit .-Partial atomic charges:...
for individual atoms, and equilibrium values of bond length
Bond length
- Explanation :Bond length is related to bond order, when more electrons participate in bond formation the bond will get shorter. Bond length is also inversely related to bond strength and the bond dissociation energy, as a stronger bond will be shorter...
s, bond angles, and dihedral angle
Dihedral angle
In geometry, a dihedral or torsion angle is the angle between two planes.The dihedral angle of two planes can be seen by looking at the planes "edge on", i.e., along their line of intersection...
s for pairs, triplets, and quadruplets of bonded atoms, and values corresponding to the effective spring constant for each potential. Most current force fields use a "fixed-charge" model by which each atom is assigned a single value for the atomic charge that is not affected by the local electrostatic environment; proposed developments in next-generation force fields incorporate models for polarizability
Polarizability
Polarizability is the measure of the change in a molecule's electron distribution in response to an applied electric field, which can also be induced by electric interactions with solvents or ionic reagents. It is a property of matter...
, in which a particle's charge is influenced by electrostatic interactions with its neighbors. For example, polarizability can be approximated by the introduction of induced dipoles; it can also be represented by Drude particle
Drude particle
Drude particles are model oscillators used to simulate the effects of electronic polarizability in the context of a classical molecular mechanics force field. It is based on the Drude model of mobile electrons...
s, or massless, charge-carrying virtual sites attached by a springlike harmonic potential
Harmonic oscillator
In classical mechanics, a harmonic oscillator is a system that, when displaced from its equilibrium position, experiences a restoring force, F, proportional to the displacement, x: \vec F = -k \vec x \, where k is a positive constant....
to each polarizable atom. The introduction of polarizability into force fields in common use has been inhibited by the high computational expense associated with calculating the local electrostatic field.
Although many molecular simulations involve biological macromolecule
Macromolecule
A macromolecule is a very large molecule commonly created by some form of polymerization. In biochemistry, the term is applied to the four conventional biopolymers , as well as non-polymeric molecules with large molecular mass such as macrocycles...
s such as protein
Protein
Proteins are biochemical compounds consisting of one or more polypeptides typically folded into a globular or fibrous form, facilitating a biological function. A polypeptide is a single linear polymer chain of amino acids bonded together by peptide bonds between the carboxyl and amino groups of...
s, DNA
DNA
Deoxyribonucleic acid is a nucleic acid that contains the genetic instructions used in the development and functioning of all known living organisms . The DNA segments that carry this genetic information are called genes, but other DNA sequences have structural purposes, or are involved in...
, and RNA
RNA
Ribonucleic acid , or RNA, is one of the three major macromolecules that are essential for all known forms of life....
, the parameters for given atom types are generally derived from observations on small organic
Organic compound
An organic compound is any member of a large class of gaseous, liquid, or solid chemical compounds whose molecules contain carbon. For historical reasons discussed below, a few types of carbon-containing compounds such as carbides, carbonates, simple oxides of carbon, and cyanides, as well as the...
molecules that are more tractable for experimental studies and quantum calculations. Different force fields can be derived from dissimilar types of experimental data, such as enthalpy
Enthalpy
Enthalpy is a measure of the total energy of a thermodynamic system. It includes the internal energy, which is the energy required to create a system, and the amount of energy required to make room for it by displacing its environment and establishing its volume and pressure.Enthalpy is a...
of vaporization
Vaporization
Vaporization of an element or compound is a phase transition from the liquid or solid phase to gas phase. There are three types of vaporization: evaporation, boiling and sublimation....
(OPLS
OPLS
The OPLS force field was developed by Prof. William L. Jorgensen at Purdue University and later at Yale University.-Functional form:The functional form of the OPLS force field is very similar to that of AMBER:...
), enthalpy
Enthalpy
Enthalpy is a measure of the total energy of a thermodynamic system. It includes the internal energy, which is the energy required to create a system, and the amount of energy required to make room for it by displacing its environment and establishing its volume and pressure.Enthalpy is a...
of sublimation (CFF
CFF
CFF may refer to:* Chemins de fer fédéraux suisses, the Swiss Federal Railways in French, the Swiss national railway.* Cambodian Freedom Fighters, a militant rebel group.* Celebrity Family Feud, a 2008 NBC game show hosted by Al Roker....
), dipole moments, or various spectroscopic parameters (CFF).
Parameter sets and functional forms are defined by force field developers to be self-consistent. Because the functional forms of the potential terms vary extensively between even closely related force fields (or successive versions of the same force field), the parameters from one force field should never be used in conjunction with the potential from another.
Deficiencies
All force fields are based on numerous approximations and derived from different types of experimental data. Therefore they are called empirical. Some existing force fields do not account for electronic polarization of the environment, an effect that can significantly reduce electrostatic interactions of partial atomic charges. This problem was addressed by developing “polarizable force fields” or using macroscopic dielectric constantDielectric constant
The relative permittivity of a material under given conditions reflects the extent to which it concentrates electrostatic lines of flux. In technical terms, it is the ratio of the amount of electrical energy stored in a material by an applied voltage, relative to that stored in a vacuum...
. However, application of a single value of dielectric constant
Dielectric constant
The relative permittivity of a material under given conditions reflects the extent to which it concentrates electrostatic lines of flux. In technical terms, it is the ratio of the amount of electrical energy stored in a material by an applied voltage, relative to that stored in a vacuum...
is questionable in the highly heterogeneous environments of proteins or biological membranes, and the nature of the dielectric depends on the model used .
All types of van der Waals force
Van der Waals force
In physical chemistry, the van der Waals force , named after Dutch scientist Johannes Diderik van der Waals, is the sum of the attractive or repulsive forces between molecules other than those due to covalent bonds or to the electrostatic interaction of ions with one another or with neutral...
s are also strongly environment-dependent, because these forces originate from interactions of induced and “instantaneous” dipoles (see Intermolecular force
Intermolecular force
Intermolecular forces are forces of attraction or repulsion which act between neighboring particles: atoms, molecules or ions. They are weak compared to the intramolecular forces, the forces which keep a molecule together...
). The original Fritz London
Fritz London
Fritz Wolfgang London was a German theoretical physicist. His fundamental contributions to the theories of chemical bonding and of intermolecular forces are today considered classic and are discussed in standard textbooks of physical chemistry.With his brother Heinz, he made a significant...
theory of these forces can only be applied in vacuum. A more general theory of van der Waals force
Van der Waals force
In physical chemistry, the van der Waals force , named after Dutch scientist Johannes Diderik van der Waals, is the sum of the attractive or repulsive forces between molecules other than those due to covalent bonds or to the electrostatic interaction of ions with one another or with neutral...
s in condensed media was developed by A. D. McLachlan in 1963 (this theory includes the original London’s approach as a special case) . The McLachlan theory predicts that van der Waals attractions in media are weaker than in vacuum and follow the "like dissolves like" rule, which means that different types of atoms interact more weakly than identical types of atoms.. This is in contrast to “combinatorial rules” or Slater-Kirkwood equation applied for development of the classical force fields. The “combinatorial rules” state that interaction energy of two dissimilar atoms (e.g. C…N) is an average of the interaction energies of corresponding identical atom pairs (i.e. C…C and N…N). According to McLachlan theory, the interactions of particles in a media can even be completely repulsive, as observed for liquid helium
Helium
Helium is the chemical element with atomic number 2 and an atomic weight of 4.002602, which is represented by the symbol He. It is a colorless, odorless, tasteless, non-toxic, inert, monatomic gas that heads the noble gas group in the periodic table...
. The conclusions of McLachlan theory are supported by direct measurements of attraction forces between different materials (Hamaker constant
Hamaker Constant
The Hamaker constant A can be defined for a Van der Waals body-body interaction:A=\pi^2\times C \times \rho_1 \times \rho_2where \rho_1 and \rho_2 are the number of atoms per unit volume in two interacting bodies and C is the coefficient in the particle-particle pair interaction.The Hamaker...
), as explained by Jacob Israelachvili
Jacob Israelachvili
Jacob Israelachvili is a professor of chemical engineering and materials at the University of California, Santa Barbara . Israelachvili received his Ph.D. in Physics from Christ's College, Cambridge in 1972, and joined UCSB in 1986. His research has involved study of molecular and interfacial forces...
in his book "Intermolecular and surface forces". It was concluded that "the interaction between hydrocarbons across water is about 10% of that across vacuum" . Such effects are unaccounted in the standard molecular mechanics.
Another round of criticism came from practical applications, such as protein structure refinement. It was noted that CASP
CASP
CASP, which stands for Critical Assessment of Techniques for Protein Structure Prediction, is a community-wide, worldwide experiment for protein structure prediction taking place every two years since 1994...
participants did not try to refine their models to avoid "a central embarrassment of molecular mechanics, namely that energy minimization or molecular dynamics generally leads to a model that is less like the experimental structure". Actually, the force fields have been successfully applied for protein structure refinement in different X-ray crystallography
X-ray crystallography
X-ray crystallography is a method of determining the arrangement of atoms within a crystal, in which a beam of X-rays strikes a crystal and causes the beam of light to spread into many specific directions. From the angles and intensities of these diffracted beams, a crystallographer can produce a...
and NMR spectroscopy
NMR spectroscopy
Nuclear magnetic resonance spectroscopy, most commonly known as NMR spectroscopy, is a research technique that exploits the magnetic properties of certain atomic nuclei to determine physical and chemical properties of atoms or the molecules in which they are contained...
applications, especially using program XPLOR . However, such refinement is driven primarily by a set of experimental constraints, whereas the force fields serve merely to remove interatomic hindrances. The results of calculations are practically the same with rigid sphere potentials implemented in program DYANA (calculations from NMR data), or with programs for crystallographic refinement that do not use any energy functions. The deficiencies of the force fields remain a major bottleneck in homology modeling
Homology modeling
Homology modeling, also known as comparative modeling of protein refers to constructing an atomic-resolution model of the "target" protein from its amino acid sequence and an experimental three-dimensional structure of a related homologous protein...
of proteins . Such situation gave rise to development of alternative empirical scoring functions specifically for ligand docking , protein folding
Protein folding
Protein folding is the process by which a protein structure assumes its functional shape or conformation. It is the physical process by which a polypeptide folds into its characteristic and functional three-dimensional structure from random coil....
, homology model refinement, computational protein design
Protein design
Protein design is the design of new protein molecules, either from scratch or by making calculated variations on a known structure. The use of rational design techniques for proteins is a major aspect of protein engineering....
, and modeling of proteins in membranes .
There is also an opinion that molecular mechanics may operate with energy which is irrelevant to protein folding or ligand binding . The parameters of typical force fields reproduce enthalpy
Enthalpy
Enthalpy is a measure of the total energy of a thermodynamic system. It includes the internal energy, which is the energy required to create a system, and the amount of energy required to make room for it by displacing its environment and establishing its volume and pressure.Enthalpy is a...
of sublimation
Sublimation (physics)
Sublimation is the process of transition of a substance from the solid phase to the gas phase without passing through an intermediate liquid phase...
, i.e. energy of evaporation of molecular crystals. However, it was recognized that protein folding
Protein folding
Protein folding is the process by which a protein structure assumes its functional shape or conformation. It is the physical process by which a polypeptide folds into its characteristic and functional three-dimensional structure from random coil....
and ligand binding are thermodynamically very similar to crystallization
Crystallization
Crystallization is the process of formation of solid crystals precipitating from a solution, melt or more rarely deposited directly from a gas. Crystallization is also a chemical solid–liquid separation technique, in which mass transfer of a solute from the liquid solution to a pure solid...
, or liquid-solid transitions, because all these processes represent “freezing” of mobile molecules in condensed media . Therefore, free energy changes during protein folding or ligand binding are expected to represent a combination of an energy similar to heat of fusion (energy absorbed during melting of molecular crystals), a conformational entropy
Conformational entropy
Conformational entropy is the entropy associated with the physical arrangement of a polymer chain that assumes a compact or globular state in solution. The concept is most commonly applied to biological macromolecules such as proteins and RNA, but can also be used for polysaccharides and other...
contribution, and solvation
Solvation
Solvation, also sometimes called dissolution, is the process of attraction and association of molecules of a solvent with molecules or ions of a solute...
free energy. The heat of fusion is significantly smaller than enthalpy of sublimation . Hence, the potentials describing protein folding or ligand binding must be weaker than potentials in molecular mechanics. Indeed, the energies of H-bonds in proteins are ~ -1.5 kcal/mol when estimated from protein engineering
Protein engineering
Protein engineering is the process of developing useful or valuable proteins. It is a young discipline, with much research taking place into the understanding of protein folding and recognition for protein design principles....
or alpha helix
Alpha helix
A common motif in the secondary structure of proteins, the alpha helix is a right-handed coiled or spiral conformation, in which every backbone N-H group donates a hydrogen bond to the backbone C=O group of the amino acid four residues earlier...
to coil
Random coil
A random coil is a polymer conformation where the monomer subunits are oriented randomly while still being bonded to adjacent units. It is not one specific shape, but a statistical distribution of shapes for all the chains in a population of macromolecules...
transition data , but the same energies estimated from sublimation
Sublimation (physics)
Sublimation is the process of transition of a substance from the solid phase to the gas phase without passing through an intermediate liquid phase...
enthalpy
Enthalpy
Enthalpy is a measure of the total energy of a thermodynamic system. It includes the internal energy, which is the energy required to create a system, and the amount of energy required to make room for it by displacing its environment and establishing its volume and pressure.Enthalpy is a...
of molecular crystal
Crystal
A crystal or crystalline solid is a solid material whose constituent atoms, molecules, or ions are arranged in an orderly repeating pattern extending in all three spatial dimensions. The scientific study of crystals and crystal formation is known as crystallography...
s were -4 to -6 kcal/mol . The depths of modified Lennard-Jones potential
Lennard-Jones potential
The Lennard-Jones potential is a mathematically simple model that approximates the interaction between a pair of neutral atoms or molecules. A form of the potential was first proposed in 1924 by John Lennard-Jones...
s derived from protein engineering data were also smaller than in typical force fields and followed the “like dissolves like” rule, as predicted by McLachlan theory .
Future Perspectives
Molecular mechanics or force field was first introduced apparently independently by Hill and by Westheimer in 1949, primarily applied to organic chemistry to estimate properties such as strain energies among others. The functional form of the force field, focused in this article applied to biological systems, was established by Lifson in the 1960s. For over a half century, force fields have served us well, providing useful insights into and interpretation of biomolecular structure and function. Undoubtedly, it will continue to be widely used, thanks to its computational efficiency, while its reliability will continue to be improved. Yet, there are many well-known deficiencies as noted above. In addition, the number of energy terms used in a given force field cannot be uniquely determined and a highly redundant number of degrees of freedom are typically used. Consequently, the "parameters" in different force fields can be vastly different. Of course, the emphasis to incorporate polarization into the standard pair-wise potentials can be very useful; however, there is no unique way of treating polarization in molecular mechanics because it is of quantum mechanical origin. Furthermore, often we are more interested in the properties derived from the dynamic dependence of the force field itself on molecular fluctuations.One possibility is that the future development of force field ought to move beyond the current molecular mechanics approach, by using quantum mechanics explicitly to construct the force field. A number of the "polarizable force fields" listed below, such as density fitting and bond-polarization, already included some of the key ingredients towards this goal. The explicit polarization (X-Pol) method appears to have established the fundamental theoretical framework for a quantal force field; the next step is to develop the necessary parameters to achieve more accurate results than classical mechanics can offer.
Popular force fields
Different force fields are designed for different purposes.MM2 was developed by Norman Allinger
Norman Allinger
Norman "Lou" Allinger is an American computational chemist and Distinguished Research Professor Emeritus of Chemistry at the University of Georgia in Athens....
primarily for conformational analysis of hydrocarbon
Hydrocarbon
In organic chemistry, a hydrocarbon is an organic compound consisting entirely of hydrogen and carbon. Hydrocarbons from which one hydrogen atom has been removed are functional groups, called hydrocarbyls....
s and other small organic molecules. It is designed to reproduce the equilibrium covalent geometry of molecules as precisely as possible. It implements a large set of parameters that is continuously refined and updated for many different classes of organic compounds (MM3 and MM4).
CFF was developed by Warshel, Lifson and coworkers as a general method for unifying studies of energies, structures and vibration of general molecules and molecular crystals. The CFF program, developed by Levitt and Warshel, is based on the Cartesian representation of all the atoms, and it served as the basis for many subsequent simulation programs.
ECEPP was developed specifically for modeling of peptides and proteins. It uses fixed geometries of amino acid residues to simplify the potential energy surface. Thus, the energy minimization is conducted in the space of protein torsion angles. Both MM2 and ECEPP include potentials for H-bonds and torsion potentials for describing rotations around single bonds. ECEPP/3 was implemented (with some modifications) in Internal Coordinate Mechanics
Internal Coordinate Mechanics
ICM stands for Internal Coordinate Mechanics and was first designed and built to predict low energy conformations of biomolecules. ICM also is a programming environment for various tasks in computational chemistry and computational structural biology, sequence analysis and rational drug design...
and FANTOM .
AMBER
AMBER
AMBER is a family of force fields for molecular dynamics of biomolecules originally developed by the late Peter Kollman's group at the University of California, San Francisco. AMBER is also the name for the molecular dynamics software package that simulates these force fields...
, CHARMM
CHARMM
CHARMM is the name of a widely used set of force fields for molecular dynamics as well as the name for the molecular dynamics simulation and analysis package associated with them...
and GROMOS
GROMOS
GROMOS is a force field for molecular dynamics simulation developed at the University of Groningen and at at the at the ETH Zurich.The united atom force field was optimized with respect to the condensed phase properties of alkanes....
have been developed primarily for molecular dynamics
Molecular dynamics
Molecular dynamics is a computer simulation of physical movements of atoms and molecules. The atoms and molecules are allowed to interact for a period of time, giving a view of the motion of the atoms...
of macromolecules, although they are also commonly applied for energy minimization. Therefore, the coordinates of all atoms are considered as free variables.
Classical force fields
- AMBERAMBERAMBER is a family of force fields for molecular dynamics of biomolecules originally developed by the late Peter Kollman's group at the University of California, San Francisco. AMBER is also the name for the molecular dynamics software package that simulates these force fields...
(Assisted Model Building and Energy Refinement) - widely used for proteins and DNA. - CHARMMCHARMMCHARMM is the name of a widely used set of force fields for molecular dynamics as well as the name for the molecular dynamics simulation and analysis package associated with them...
(Chemistry at HARvard Molecular Mechanics) - originally developed at Harvard, widely used for both small molecules and macromolecules - CHARMmCHARMMCHARMM is the name of a widely used set of force fields for molecular dynamics as well as the name for the molecular dynamics simulation and analysis package associated with them...
- commercial version of CHARMM, available through AccelrysAccelrysAccelrys is a software company headquartered in the US, with representation in Europe and Japan. It provides software for chemical, materials and bioscience research for the pharmaceutical, biotechnology, consumer packaged goods, aerospace, energy and chemical industries.Accelrys started in 2001...
. - CVFF - also broadly used for small molecules and macromolecules.
- COSMOS-NMR - hybrid QM/MM forcefield adapted to a variety of inorganic compounds, organic compounds and biological macromolecules, including semi-empirical calculation of atomic charges and NMR properties. COSMOS-NMR is optimized for NMR based structure elucidation and implemented in COSMOS molecular modelling package.
- GROMACS - the force field optimized for the molecular dynamics package of the same name.
- GROMOSGROMOSGROMOS is a force field for molecular dynamics simulation developed at the University of Groningen and at at the at the ETH Zurich.The united atom force field was optimized with respect to the condensed phase properties of alkanes....
- a force field that comes as part of the GROMOS (GROningen MOlecular Simulation package), a general-purpose molecular dynamics computer simulation package for the study of biomolecular systems. GROMOS force field (A-version) has been developed for application to aqueous or apolar solutions of proteins, nucleotides and sugars. However, a gas phase version (B-version) for simulation of isolated molecules is also available. - OPLSOPLSThe OPLS force field was developed by Prof. William L. Jorgensen at Purdue University and later at Yale University.-Functional form:The functional form of the OPLS force field is very similar to that of AMBER:...
(Optimized Potential for Liquid Simulations) (variations include OPLS-AA, OPLS-UA, OPLS-2001, OPLS-2005) - developed by William L. Jorgensen at the Yale University Department of Chemistry. - ENZYMIX – a general polarizable force field for modeling chemical reactions in biological molecules. This force field is implemented with the empirical valence bond (EVB) method and is also combined with the semimacroscopic PDLD approach in the program in the MOLARIS package.
- ECEPP/2 - first force field for polypeptide molecules - developed by F.A. Momany, H.A. Scheraga and colleagues.
- QCFF/PI – A general force field for conjugated molecules..
- UFFUniversal force fieldUniversal force field is an all atom potential containing parameters for every atom. The force field parameters are estimated using general rules based only on the element, its hybridization, and its connectivity....
- A general force field with parameters for the full periodic table up to and including the actinoids - developed at Colorado State University.
Second-generation force fields
- CFFCFFCFF may refer to:* Chemins de fer fédéraux suisses, the Swiss Federal Railways in French, the Swiss national railway.* Cambodian Freedom Fighters, a militant rebel group.* Celebrity Family Feud, a 2008 NBC game show hosted by Al Roker....
- a family of forcefields adapted to a broad variety of organic compounds, includes force fields for polymers, metals, etc. - MMFFMerck Molecular Force FieldMerck Molecular Force Field is a family of force fields developed by Merck Research Laboratories. They are based on the MM3 force field. MMFF is not optimized for a single use , but tries to perform well for a wide range of organic chemistry calculations...
(Merck Molecular Force Field)- developed at Merck, for a broad range of molecules. - MM2, MM3, MM4 - developed by Norman AllingerNorman AllingerNorman "Lou" Allinger is an American computational chemist and Distinguished Research Professor Emeritus of Chemistry at the University of Georgia in Athens....
, parametrized for a broad range of molecules. - QVBMM - developed by Vernon G. S. Box, parameterized for all biomolecules and a broad range of organic molecules, and implemented in StruMM3D (STR3DI32).
Polarizable force fields based on electronic structural theory
- X-Pol: the Explicit Polarization TheoryX-Pol: the Explicit Polarization TheoryThe Explicit Polarization Theory is both a fragment-based electronic structure method and a quantal force field for macromolecular systems.-Introduction:...
http://www.x-pol.org - a fragment-based electronic structure method introduced by Jiali Gao http://jialigao.org at the University of Minnesota, which can be used at any level of theory—ab initio Hartree-Fock (HF), semiempirical molecular orbital theory, correlated wave function theory, or Kohn-Sham (KS) density functional theory (DFT). It is capable of performing more than 3200 steps (3.2 ps) of MD simulations of a fully solvated protein in water with periodic boundary conditions, consisting of about 15000 atoms and 30000 basis functions on a single processor in 24 hours in 2008, with a full quantum mechanical representation of the entire system. Note that the first MD simulation of a protein by Gelin, McCammon and Karplus in 1979 lasted just over 9 ps using a United-Atom force field without solvent.
Polarizable force field based on induced dipole
- CFF/ind and ENZYMIX – The first polarizable force field which has subsequently been used in many applications to biological systems..
- DRF90 developed by P. Th. van Duijnen and coworkers.
- PIPF – The polarizable intermolecular potential for fluids is an induced point-dipole force field for organic liquids and biopolymers. The molecular polarization is based on Thole's interacting dipole (TID) model and was developed by Jiali Gao http://jialigao.org at the University of Minnesota.
Polarizable force fields based on point charges
- PFF (Polarizable Force Field) developed by Richard A. Friesner and coworkers.
- SP-basis Chemical Potential Equalization (CPE) approach developed by R. Chelli and P. Procacci.
- CHARMM polarizable force field developed by S. Patel (University of Delaware) and C. L. Brooks III (University of Michigan).
- AMBER polarizable force field developed by Jim Caldwell and coworkers.
- CHARMM polarizable force field based on the classical Drude oscillator developed by A. MacKerell (University of Maryland, Baltimore) and B. Roux (University of Chicago).
Polarizable force fields based on distributed multipoles
- The SIBFA (Sum of Interactions Between Fragments Ab initio computed) force field for small molecules and flexible proteins, developed by Nohad Gresh (Paris V, René Descartes University) and Jean-Philip Piquemal (Paris VI, Pierre & Marie Curie University). SIBFA is a molecular mechanics procedure formulated and calibrated on the basis of ab initio supermolecule computations. Its purpose is to enable the simultaneous and reliable computations of both intermolecular and conformational energies governing the binding specificities of biologically and pharmacologically relevant molecules. This procedure enables an accurate treatment of transition metals. The inclusion of a ligand field contribution allows computations on "open-shell" metalloproteins.
- AMOEBA (Atomic Multipole Optimized Energetics for Biomolecular Applications) force field developed by Pengyu Ren (University of Texas at Austin) and Jay W. Ponder (Washington University).
- ORIENT procedure developed by Anthony J. Stone (Cambridge University) and coworkers.
- Non-Empirical Molecular Orbital (NEMO) procedure developed by Gunnar Karlström and coworkers at Lund University (Sweden)
Polarizable force fields based on density
- Gaussian Electrostatic Model (GEM) - a polarizable force field based on Density Fitting developed by Thomas A. Darden and G. Andrés Cisneros at NIEHS; and Jean-Philip Piquemal (Paris VI University).
- Polarizable procedure based on the Kim-Gordon approach developed by Jürg Hutter and coworkers (University of Zürich)
Polarizable force fields based on Bond Polarization Theory (BPT)
- COSMOS-NMR (Computer Simulation of Molecular Structure) - developed by Ulrich Sternberg and coworkers. Hybrid QM/MM force field enables explicit quantum-mechanical calculation of electrostatic properties using localized bond orbitals with fast BPT formalism. Atomic charge fluctuation is possible in each molecular dynamics step.
Reactive force fields
- ReaxFFReaxFFReaxFF is a force field developed by Adri van Duin, William A. Goddard, III and co-workers at the California Institute of Technology for use e.g. in molecular dynamics simulations...
- reactive force field developed by Adri van Duin, William Goddard and coworkers. It is fast, transferable and is the computational method of choice for atomistic-scale dynamical simulations of chemical reactions. - EVB (empirical valence bond) - this reactive force field, introduced by Warshel and coworkers, is probably the most reliable and physically consistent way of using force fields in modeling chemical reactions in different environments. The EVB facilitates calculations of actual activation free energies in condensed phases and in enzymes.
- RWFF - reactive force field for water developed by Detlef W. M. Hofmann, Liudmila N. Kuleshova and Bruno D'Aguanno. It is very fast, reproduces the experimental data of neutron scattering accurately, and allows the simulation of bond formation/breaking of water and acids.
Coarse-grained force fields
- VAMM (Virtual atom molecular mechanics) - a coarse-grained force field developed by Korkut and Hendrickson for molecular mechanics calculations such as large scale conformational transitions based on the virtual interactions of C-alpha atoms. It is a knowledge based force field and formulated to capture features dependent on secondary structure and on residue-specific contact information in proteins.
Other
- VALBONDVALBONDIn molecular mechanics, VALBOND is a method for computing the angle bending energy that is based on valence bond theory. It is based on orbital strength functions, which are maximized when the hybrid orbitals on the atom are orthogonal...
- a function for angle bending that is based on valence bond theoryValence bond theoryIn chemistry, valence bond theory is one of two basic theories, along with molecular orbital theory, that were developed to use the methods of quantum mechanics to explain chemical bonding. It focuses on how the atomic orbitals of the dissociated atoms combine to give individual chemical bonds...
and works for large angular distortions, hypervalent moleculeHypervalent moleculeA hypervalent molecule is a molecule that contains one or more main group elements formally bearing more than eight electrons in their valence shells...
s, and transition metal complexesComplex (chemistry)In chemistry, a coordination complex or metal complex, is an atom or ion , bonded to a surrounding array of molecules or anions, that are in turn known as ligands or complexing agents...
. It can be incorporated into other force fields such as CHARMM and UFF.
Water models
The set of parameters used to model water or aqueous solutions (basically a force field for water) is called a water modelWater model
In computational chemistry, classical water models are used for the simulation of water clusters, liquid water, and aqueous solutions with explicit solvent. These models use the approximations of molecular mechanics...
. Water has attracted a great deal of attention due to its unusual properties and its importance as a solvent. Many water models have been proposed; some examples are TIP3P, TIP4P, SPC, Flexible SPC
Flexible SPC water model
The Flexible Simple Point Charge water model is a re-parametrization of the three-site SPC water model. The SPC model is rigid, whilst the flexible SPC model is flexible. In the model of Toukan and Rahman, the O-H stretching is made anharmonic and thus the dynamical behavior is well described...
, and ST2.
See also
Further reading
- Schlick T. (2000). Molecular Modeling and Simulation: An Interdisciplinary Guide Interdisciplinary Applied Mathematics: Mathematical Biology. Springer-Verlag New York, NY.
- Israelachvili, J.N. (1992) Intermolecular and surface forces. Academic Press, San Diego.
- Warshel A (1991). "Computer Modeling of Chemical Reactions in Enzymes and Solutions" John Wiley & Sons New York.

