DNA nanotechnology
Encyclopedia
DNA nanotechnology is a branch of nanotechnology
which uses the molecular recognition
properties of DNA
and other nucleic acid
s to create designed, artificial structures out of DNA for technological purposes. In this field, DNA is used as a structural material rather than as a carrier of genetic information, making it an example of bionanotechnology. DNA nanotechnology has applications in molecular self-assembly
and in DNA computing
.
Although DNA is usually considered in the context of molecular biology
as the carrier of genetic information
in living cells
, DNA nanotechnology considers DNA solely as a chemical and as a material, and is usually pursued outside of any biological context. DNA nanotechnology makes use of the fact that, due to the specificity of Watson-Crick base pair
ing, only portions of the strands which are complementary
to each other will bind to each other to form duplex
DNA. DNA nanotechnology attempts to rationally design
sets of DNA strands so that desired portions of each strand will assemble in the correct positions for some desired target structure, a process called nucleic acid design
.
Although the field is usually called DNA nanotechnology, its principles apply equally well to other nucleic acids such as RNA
and PNA, and structures incorporating these have been made. For this reason the field is occasionally referred to as nucleic acid nanotechnology.
ing in nucleic acid molecules. The structure
of a nucleic acid molecule consists of a sequence of nucleotide
s, distinguished by which nucleobase
they contain. In DNA, the four bases used are adenine
(A), cytosine
(C), guanine
(G), and thymine
(T). Nucleic acids have the property that two molecules will bind to each other to form a double helix only if the two sequences are complementary
, meaning that they form matching sequences of base pairs, with A's only binding to T's, and C's only to G's. Because the formation of correctly matched base pairs is energetically favorable
, nucleic acid strands are expected in most cases to bind to each other in the conformation that maximizes the number of correctly paired bases. This property, that the sequence determines the pattern of binding and the overall structure, is used by the field of DNA nanotechnology in that sequences are artificially designed
so that a desired structure is favored to form.
Nearly all structures in DNA nanotechnology make use of branched DNA structures containing junctions, as opposed to most biological DNA which exists in a linear double helix
form. One of the simplest branched structures, and the first made, is a four-arm junction which can be made using four individual DNA strands which are complementary to each other in the correct pattern. Unlike in natural Holliday junction
s, in the artificial immobile four-arm junction shown below, the base sequence of each arm is different, meaning that the junction point is fixed in a certain position.
Junctions can be used in more complex molecules. One of the more widely used of these is the "double-crossover" or DX motif. A DX molecule can be thought of as two DNA duplexes positioned parallel to each other, with two crossover points where strands cross from one duplex into the other. Each junction point is itself topologically a four-arm junction. This molecule has the advantage that the junction points are now constrained to a single orientation as opposed to being flexible as in the four-arm junction. This makes the DX motif suitable as a structural building block for larger DNA complexes.
so that the individual nucleic acid strands will assemble into the desired structures. The design process of such nanostructures usually begins with the specification of a desired target structure and/or functionality. Then, the overall secondary structure
of the target molecule is designed, meaning the arrangement of nucleic acid strands within the structure, and which portions of those strands should be bound to each other. The last step is the primary structure design, the specification of the actual base sequences of each nucleic acid strand.
, or the series of base pairs which hold the individual strands together in the desired shape. There are several approaches which have been demonstrated:
sequence to each strand so that they will associate into a desired conformation. Nucleic acid design is central to the field of DNA nanotechnology. Most methods seek to designing sequences so that the target structure is a thermodynamic minimum, and mis-assembled structures have higher energies and are thus disfavored. This is done either through heuristic methods such as sequence symmetry minimization and coding theory based approaches, or by explicitly using a full nearest-neighbor thermodynamic model. Geometric models are also used to examine tertiary structure
of the nanostructures and ensure that the complexes are not overly strained
.
Nucleic acid design has similar goals to protein design
: in both, the sequence of monomers is designed to favor the desired folded or associated structure and to disfavor alternate structures. Nucleic acid design has the advantage of being a much computationally simpler problem, since the simplicity of Watson-Crick base pair
ing rules leads to simple heuristic
methods which yield experimentally robust designs. However, nucleic acid structures are less versatile than proteins in their functionality.
— has a robust, defined geometry which makes it possible to predict and design the structures of more complex DNA molecules. Many such structures have been created, including two- and three-dimensional structures; and periodic, aperiodic, and discrete structures.
 Smaller nucleic acid assemblies can be equipped with sticky ends in order to combine them into a two-dimensional periodic lattice. The earliest example of this was the array of DX, or double-crossover, molecules. Each DX molecule can be designed with four sticky ends, one at each end of the two double-helical
Smaller nucleic acid assemblies can be equipped with sticky ends in order to combine them into a two-dimensional periodic lattice. The earliest example of this was the array of DX, or double-crossover, molecules. Each DX molecule can be designed with four sticky ends, one at each end of the two double-helical
domains, and these sticky ends can be designed with sequences that cause the DX units to combine into a specific tessellated
pattern. They thus form extended flat sheets which are essentially rigid two-dimensional crystal
s of DNA.
Two-dimensional arrays have been made out of other motifs as well, including the Holliday junction
rhombus
array
as well as various DX-based arrays making use of a double-cohesion scheme.
Creating three-dimensional lattices out of DNA was the earliest goal of DNA nanotechnology, but proved to be one of the most difficult to realize. Success in constructing three-dimensional DNA lattices was finally reported in 2009 using a motif based on the concept of tensegrity
, a balance between tension and compression forces.
s, but the carbon nanotubes are stronger and better conductors, whereas the DNA nanotubes are more easily modified and connected to other structures. There have been multiple schemes for constructing DNA nanotubes, one of which uses the inherent curvature of DX tiles to form a DX lattice to curl around itself and close into a tube. An alternative design uses single-stranded "tiles" for which the rigidity of the tube is an emergent property. This method also has the benefit of being able to determine the circumference of the nanotube in a simple, modular fashion.
such as an octahedron or cube. In other words, the DNA duplexes trace the edges of a polyhedron with a DNA junction at each vertex. The earliest demonstrations of DNA polyhedra involved multiple ligations
and solid-phase synthesis
steps to create catenated
polyhedra.
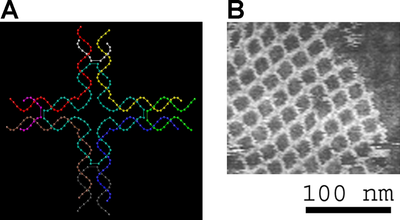
More recent work has yielded polyhedra whose synthesis is much easier. These include a DNA octahedron
made from a long single strand designed to fold into the correct conformation, as well as a tetrahedron which can be produced from four DNA strands in a single step, pictured at the top of this article.
, as most other DNA nanotechnology methods do. DNA origami was first demonstrated for two-dimensional shapes; demonstrated designs included the smiley face and a coarse map
of North America
. This was later extended to solid three-dimensional shapes.
In addition, structures have been constructed with two-dimensional faces which fold into an overall three-dimensional shape, akin to a cardboard box. These can be programmed to open and release their cargo in response to a stimulus, making them potentially useful as programmable molecular cages
.
but only recently has progress been made in reducing these kinds of schemes to practice. In 2006, researchers covalently attached gold nanoparticles to a DX-based tile and showed that self-assembly of the DNA structures also assembled the nanoparticles hosted on them.
Also that year, Dwyer and LaBean demonstrated the letters "D" "N" and "A" created on a 4x4 DX array using streptavidin, and a hierarchical assembly based on this approach was also demonstrated that scales to larger arrays (8X8 and 8.96 MD).
A non-covalent hosting scheme was shown in 2007, using Dervan
polyamides on a DX array to arrange streptavidin
proteins on specific kinds of tiles on the DNA array.
There has also been interest in using DNA nanotechnology to assemble molecular electronics devices. To this end, DNA has been used to assemble single walled carbon nanotubes into field-effect transistor
s.

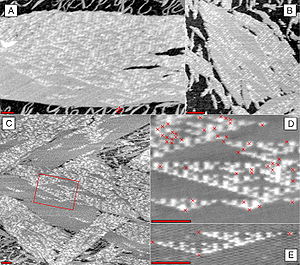
DNA nanotechnology has been applied to the related field of DNA computing. The DX tiles can have their sticky end sequences chosen so that they act as Wang tile
s, allowing them to perform computation. A DX array has been demonstrated whose assembly encodes an XOR operation; this allows the DNA array to implement a cellular automaton
which generates a fractal
called the Sierpinski gasket. Another system has the function of a binary counter
, displaying a representation of increasing binary numbers as it grows. These results show that computation can be incorporated into the assembly of DNA arrays, increasing its scope beyond simple periodic arrays.
Note that DNA computing overlaps with, but is distinct from, DNA nanotechnology. The latter uses the specificity of Watson-Crick basepairing to make novel structures out of DNA. These structures can be used for DNA computing, but they do not have to be. Additionally, DNA computing can be realized without using the types of molecules made possible by DNA nanotechnology.
. DNA machines have also been made which show a twisting motion. The first such device made use of the transition between the B-DNA and Z-DNA
forms to respond to a change in buffer conditions.
This reliance on buffer conditions, however, caused all devices to change state at the same time. A subsequent system, called "molecular tweezers," changes from an open to a closed state based upon the presence of control strands, allowing multiple devices to be individually operated in solution. This was followed up by another system which relies on the presence of control strands to switch from a paranemic-crossover (PX) conformation to a double-junction (JX2) conformation.
Nucleic acid nanomachines have been made which exhibit directional motion along a linear track, called DNA walkers. A large number of schemes have been demonstrated. One strategy is to control the motion of the walker along the track using control strands which need to be manually added in sequence. Another approach is to make use of restriction enzyme
s or deoxyribozyme
s to cleave the strands and cause the walker to move forward, which has the advantage of running autonomously. A later system extended the concept of DNA walkers to walk upon a two-dimensional surface rather than a linear track, and demonstrated the ability to selectively pick up and move molecular cargo. Additionally, a linear walker has been demonstrated which performs DNA-templated synthesis
as the walker advances along the track, allowing autonomous multistep chemical synthesis directed by the walker.
and biophysics
. One such application, the earliest envisaged for the field, is in crystallography
, where molecules which are hard to crystallize by themselves could be arranged and oriented within a three-dimensional nucleic acid lattice, thus allowing determination of their structure. DNA origami rods have also been used to replace liquid crystals in residual dipolar coupling
experiments in protein NMR spectroscopy; using DNA origami is advantageous because, unlike liquid crystals, they are tolerant of the detergents needed to suspend membrane protein
s in solution. It has been demonstrated that DNA walkers can be used as nanoscale assembly line
s to move nanoparticle
s and direct chemical synthesis
. Furthermore, DNA origami structures have aided in the biophysical studies of enzyme function and protein folding
.
DNA nanotechnology is also moving towards potential real-world applications. It has been suggested that the ability of nucleic acid arrays to arrange other molecules has potential applications in molecular scale electronics, with the assembly of a nucleic acid lattice templating the assembly of molecular electronic elements such as molecular wires. DNA nanotechnology has also been called a form of programmable matter
because of the coupling of computation to its material properties.
There are also potential applications for DNA nanotechnology in nanomedicine
, making use of its ability to perform computation in a biocompatible
format to make "smart drugs" for targeted drug delivery
. One such system being investigated uses a hollow DNA box containing proteins which induce apoptosis
, or cell death, which will only open when in proximity to a cancer cell
.
software. Once the sequences have been designed, the nucleic acid molecules themselves are synthesized through standard oligonucleotide synthesis
methods. This process is usually automated by using a machine called an oligonucleotide synthesizer, and nucleic acids of custom sequence are commercially available from many vendors. For methods which require pure strands of known concentration, the nucleic acid strands can be purified using denaturing gel electrophoresis, and concentrations are determined by one of several nucleic acid quantitation methods using ultraviolet absorbance spectroscopy.
The fully formed target structures are usually characterized by native gel electrophoresis, which provides information about the size and shape of DNA molecules. An electrophoretic mobility shift assay
can indicate whether a stucture incorporates all the individual desired strands. Fluorescent labeling and Förster resonance energy transfer (FRET) are also used to characterize the structure of the molecules.
Nucleic acid structures can be directly imaged by atomic force microscopy, which is well-suited to extended two-dimensional structures, but is less useful for discrete three-dimensional structures due to the interaction of the microscope tip with the fragile nucleic acid structure. For these latter structures transmission electron microscopy
and cryo-electron microscopy
are important methods. Extended three-dimensional lattices are analyzed by X-ray crystallography
.
in the early 1980s. Seeman was originally concerned with using a three-dimensional DNA lattice to orient target molecules, which would simplify their crystallographic study
by eliminating the difficult process of obtaining pure crystals. This idea had reportedly come to him in fall 1980, after realizing the similarity between the M. C. Escher
woodcut Depth and an array of DNA six-arm junctions.
To this end, Seeman's laboratory in 1991 published the synthesis of a cube
made of DNA, the first three-dimensional nanoscale object, for which he received the 1995 Feynman Prize in Nanotechnology
, which was followed by a DNA truncated octahedron
. However, it soon became clear that these molecules, polygonal shapes with flexible junctions as their vertices
, were not rigid enough to form extended three-dimensional lattices. Seeman developed the more rigid double-crossover (DX) motif
, and in collaboration with Erik Winfree
, in 1998 published the creation of two-dimensional lattices of DX tiles. The synthesis of a three-dimensional lattice was finally published by Seeman in 2009, nearly thirty years after he had set out to do so.
These tile-based structures had the advantage that they provided the capability to implement DNA computing
, which was demonstrated by Winfree and Paul Rothemund in their 2004 paper on the algorithmic self-assembly of a Sierpinski gasket structure, and for which they shared the 2006 Feynman Prize in Nanotechnology. Winfree's key insight was that the DX tiles could be used as Wang tile
s, meaning that their assembly was capable of performing computation.
The first DNA nanomachine
—a motif which changes its structure in response to an input—was demonstrated in 1999 by Seeman. An improved system was demonstrated by Bernard Yurke the following year, which was also the first nucleic acid device to make use of toehold-mediated strand displacement. The next advance was to translate this into mechanical motion, and in 2004 and 2005, a number of DNA walker systems were demonstrated by the groups of Seeman, Niles Pierce, Andrew Turberfield
, and Chengde Mao.
In 2006, Rothemund first demonstrated the new DNA origami
technique for easily and robustly creating folded DNA molecules of any shape. Rothemund had conceived of this method as being conceptually intermediate between Seeman's DX lattices, which used many short strands, and William Shih's DNA octahedron, which consisted mostly of one very long strand; Rothemund's DNA origami contains a long strand whose folding is assisted by a number of short strands. This method allowed the creation of much larger structures than were previously possible, which are less technically demanding to design and synthesize. DNA origami was the cover story of Nature
on March 15, 2006. Rothemund's research demonstrating two-dimensional DNA origami structures was followed by the demonstration of solid three-dimensional DNA origami by Shih in 2009, while the labs of Jørgen Kjems and Hao Yan demonstrated hollow three-dimensional structures made out of two-dimensional faces.
DNA nanotechnology as a field was initially met with some skepticism due to the unusual non-biological use of nucleic acids as a material for building structures and doing computation, as well as the preponderance of proof of principle experiments which extended the capabilities of the field but were far from actual applications. Seeman's seminal 1991 paper on the synthesis of the DNA cube was rejected by the journal Science
after one reviewer praised its originality while another criticized it for its lack of biological relevance. By the early 2010s, however, the field was considered to have increased its capabilities to the point that applications for basic science research were beginning to be realized, and practical applications in medicine and other fields were beginning to be considered feasible. In addition, the field had grown from very few active laboratories in 2001, to at least 60 in 2010, which increased the talent pool and thus the number of advancements in the field during that decade.
—An article written for laypeople by the founder of the field
—An overview of the results of the field from the perspective of self-assembly
—A review including some older results in the field
—A review of more recent results in the period 2001–2010
—A review coming from the viewpoint of secondary structure design
—A minireview specifically focusing on tile-based assembly
—A review of nucleic acid nanomechanical devices
—A review of DNA systems making use of strand displacement mechanisms
Nanotechnology
Nanotechnology is the study of manipulating matter on an atomic and molecular scale. Generally, nanotechnology deals with developing materials, devices, or other structures possessing at least one dimension sized from 1 to 100 nanometres...
which uses the molecular recognition
Molecular recognition
The term molecular recognition refers to the specific interaction between two or more molecules through noncovalent bonding such as hydrogen bonding, metal coordination, hydrophobic forces, van der Waals forces, π-π interactions, electrostatic and/or electromagnetic effects...
properties of DNA
DNA
Deoxyribonucleic acid is a nucleic acid that contains the genetic instructions used in the development and functioning of all known living organisms . The DNA segments that carry this genetic information are called genes, but other DNA sequences have structural purposes, or are involved in...
and other nucleic acid
Nucleic acid
Nucleic acids are biological molecules essential for life, and include DNA and RNA . Together with proteins, nucleic acids make up the most important macromolecules; each is found in abundance in all living things, where they function in encoding, transmitting and expressing genetic information...
s to create designed, artificial structures out of DNA for technological purposes. In this field, DNA is used as a structural material rather than as a carrier of genetic information, making it an example of bionanotechnology. DNA nanotechnology has applications in molecular self-assembly
Molecular self-assembly
Molecular self-assembly is the process by which molecules adopt a defined arrangement without guidance or management from an outside source. There are two types of self-assembly, intramolecular self-assembly and intermolecular self-assembly...
and in DNA computing
DNA computing
DNA computing is a form of computing which uses DNA, biochemistry and molecular biology, instead of the traditional silicon-based computer technologies. DNA computing, or, more generally, biomolecular computing, is a fast developing interdisciplinary area...
.
Although DNA is usually considered in the context of molecular biology
Molecular biology
Molecular biology is the branch of biology that deals with the molecular basis of biological activity. This field overlaps with other areas of biology and chemistry, particularly genetics and biochemistry...
as the carrier of genetic information
Genetics
Genetics , a discipline of biology, is the science of genes, heredity, and variation in living organisms....
in living cells
Cell (biology)
The cell is the basic structural and functional unit of all known living organisms. It is the smallest unit of life that is classified as a living thing, and is often called the building block of life. The Alberts text discusses how the "cellular building blocks" move to shape developing embryos....
, DNA nanotechnology considers DNA solely as a chemical and as a material, and is usually pursued outside of any biological context. DNA nanotechnology makes use of the fact that, due to the specificity of Watson-Crick base pair
Base pair
In molecular biology and genetics, the linking between two nitrogenous bases on opposite complementary DNA or certain types of RNA strands that are connected via hydrogen bonds is called a base pair...
ing, only portions of the strands which are complementary
Complementarity (molecular biology)
In molecular biology, complementarity is a property of double-stranded nucleic acids such as DNA, as well as DNA:RNA duplexes. Each strand is complementary to the other in that the base pairs between them are non-covalently connected via two or three hydrogen bonds...
to each other will bind to each other to form duplex
Nucleic acid double helix
In molecular biology, the term double helix refers to the structure formed by double-stranded molecules of nucleic acids such as DNA and RNA. The double helical structure of a nucleic acid complex arises as a consequence of its secondary structure, and is a fundamental component in determining its...
DNA. DNA nanotechnology attempts to rationally design
Rational design
In chemical biology and biomolecular engineering, rational design is the strategy of creating new molecules with a certain functionality, based upon the ability to predict how the molecule's structure will affect its behavior through physical models...
sets of DNA strands so that desired portions of each strand will assemble in the correct positions for some desired target structure, a process called nucleic acid design
Nucleic acid design
Nucleic acid design is the process of generating a set of nucleic acid base sequences that will associate into a desired conformation. Nucleic acid design is central to the fields of DNA nanotechnology and DNA computing...
.
Although the field is usually called DNA nanotechnology, its principles apply equally well to other nucleic acids such as RNA
RNA
Ribonucleic acid , or RNA, is one of the three major macromolecules that are essential for all known forms of life....
and PNA, and structures incorporating these have been made. For this reason the field is occasionally referred to as nucleic acid nanotechnology.
Fundamental concepts
DNA nanotechnology creates complex structures out of nucleic acids by making use of the specificity of base pairBase pair
In molecular biology and genetics, the linking between two nitrogenous bases on opposite complementary DNA or certain types of RNA strands that are connected via hydrogen bonds is called a base pair...
ing in nucleic acid molecules. The structure
Nucleic acid structure
Nucleic acid structure refers to the structure of nucleic acids such as DNA and RNA It is often divided into four different levels:* Primary structure—the raw sequence of nucleobases of each of the component DNA strands;...
of a nucleic acid molecule consists of a sequence of nucleotide
Nucleotide
Nucleotides are molecules that, when joined together, make up the structural units of RNA and DNA. In addition, nucleotides participate in cellular signaling , and are incorporated into important cofactors of enzymatic reactions...
s, distinguished by which nucleobase
Nucleobase
Nucleobases are a group of nitrogen-based molecules that are required to form nucleotides, the basic building blocks of DNA and RNA. Nucleobases provide the molecular structure necessary for the hydrogen bonding of complementary DNA and RNA strands, and are key components in the formation of stable...
they contain. In DNA, the four bases used are adenine
Adenine
Adenine is a nucleobase with a variety of roles in biochemistry including cellular respiration, in the form of both the energy-rich adenosine triphosphate and the cofactors nicotinamide adenine dinucleotide and flavin adenine dinucleotide , and protein synthesis, as a chemical component of DNA...
(A), cytosine
Cytosine
Cytosine is one of the four main bases found in DNA and RNA, along with adenine, guanine, and thymine . It is a pyrimidine derivative, with a heterocyclic aromatic ring and two substituents attached . The nucleoside of cytosine is cytidine...
(C), guanine
Guanine
Guanine is one of the four main nucleobases found in the nucleic acids DNA and RNA, the others being adenine, cytosine, and thymine . In DNA, guanine is paired with cytosine. With the formula C5H5N5O, guanine is a derivative of purine, consisting of a fused pyrimidine-imidazole ring system with...
(G), and thymine
Thymine
Thymine is one of the four nucleobases in the nucleic acid of DNA that are represented by the letters G–C–A–T. The others are adenine, guanine, and cytosine. Thymine is also known as 5-methyluracil, a pyrimidine nucleobase. As the name suggests, thymine may be derived by methylation of uracil at...
(T). Nucleic acids have the property that two molecules will bind to each other to form a double helix only if the two sequences are complementary
Complementarity (molecular biology)
In molecular biology, complementarity is a property of double-stranded nucleic acids such as DNA, as well as DNA:RNA duplexes. Each strand is complementary to the other in that the base pairs between them are non-covalently connected via two or three hydrogen bonds...
, meaning that they form matching sequences of base pairs, with A's only binding to T's, and C's only to G's. Because the formation of correctly matched base pairs is energetically favorable
Nucleic acid thermodynamics
Nucleic acid thermodynamics is the study of the thermodynamics of nucleic acid molecules, or how temperature affects nucleic acid structure. For multiple copies of DNA molecules, the melting temperature is defined as the temperature at which half of the DNA strands are in the double-helical state...
, nucleic acid strands are expected in most cases to bind to each other in the conformation that maximizes the number of correctly paired bases. This property, that the sequence determines the pattern of binding and the overall structure, is used by the field of DNA nanotechnology in that sequences are artificially designed
Nucleic acid design
Nucleic acid design is the process of generating a set of nucleic acid base sequences that will associate into a desired conformation. Nucleic acid design is central to the fields of DNA nanotechnology and DNA computing...
so that a desired structure is favored to form.
Nearly all structures in DNA nanotechnology make use of branched DNA structures containing junctions, as opposed to most biological DNA which exists in a linear double helix
Nucleic acid double helix
In molecular biology, the term double helix refers to the structure formed by double-stranded molecules of nucleic acids such as DNA and RNA. The double helical structure of a nucleic acid complex arises as a consequence of its secondary structure, and is a fundamental component in determining its...
form. One of the simplest branched structures, and the first made, is a four-arm junction which can be made using four individual DNA strands which are complementary to each other in the correct pattern. Unlike in natural Holliday junction
Holliday junction
A Holliday junction is a mobile junction between four strands of DNA. The structure is named after Robin Holliday, who proposed it in 1964 to account for a particular type of exchange of genetic information he observed in yeast known as homologous recombination...
s, in the artificial immobile four-arm junction shown below, the base sequence of each arm is different, meaning that the junction point is fixed in a certain position.
Junctions can be used in more complex molecules. One of the more widely used of these is the "double-crossover" or DX motif. A DX molecule can be thought of as two DNA duplexes positioned parallel to each other, with two crossover points where strands cross from one duplex into the other. Each junction point is itself topologically a four-arm junction. This molecule has the advantage that the junction points are now constrained to a single orientation as opposed to being flexible as in the four-arm junction. This makes the DX motif suitable as a structural building block for larger DNA complexes.
Design
DNA nanostructures must be rationally designedRational design
In chemical biology and biomolecular engineering, rational design is the strategy of creating new molecules with a certain functionality, based upon the ability to predict how the molecule's structure will affect its behavior through physical models...
so that the individual nucleic acid strands will assemble into the desired structures. The design process of such nanostructures usually begins with the specification of a desired target structure and/or functionality. Then, the overall secondary structure
Nucleic acid secondary structure
The secondary structure of a nucleic acid molecule refers to the basepairing interactions within a single molecule or set of interacting molecules, and can be represented as a list of bases which are paired in a nucleic acid molecule....
of the target molecule is designed, meaning the arrangement of nucleic acid strands within the structure, and which portions of those strands should be bound to each other. The last step is the primary structure design, the specification of the actual base sequences of each nucleic acid strand.
Structural design
The first step in designing a nucleic acid nanostructure is to decide how a given structure should be represented by a specific arrangement of nucleic acid strands. This design step thus determines the secondary structureNucleic acid secondary structure
The secondary structure of a nucleic acid molecule refers to the basepairing interactions within a single molecule or set of interacting molecules, and can be represented as a list of bases which are paired in a nucleic acid molecule....
, or the series of base pairs which hold the individual strands together in the desired shape. There are several approaches which have been demonstrated:
- Tile-based structures. This approach breaks the target structure into smaller units with strong binding between the strands contained in each unit, and relatively weaker interactions between the units. It is often used to make periodic lattices, but can also be used to implement algorithmic self-assembly, making them one platform for DNA computingDNA computingDNA computing is a form of computing which uses DNA, biochemistry and molecular biology, instead of the traditional silicon-based computer technologies. DNA computing, or, more generally, biomolecular computing, is a fast developing interdisciplinary area...
. - Folding structures. An alternative to the tile-based approach, folding approaches make the nanostructure out of a single long strand. This long strand can either have a designed sequence which folds due to its interactions with itself, or it can be folded into the desired shape by using shorter, "staple" strands. This latter method is called DNA origamiDNA origamiDNA origami is the nanoscale folding of DNA to create arbitrary two and three dimensional shapes at the nanoscale. The specificity of the interactions between complementary base pairs make DNA a useful construction material through design of its base sequences...
, which allows the creation of two- and three-dimensional shapes at the nanoscale using DNADNADeoxyribonucleic acid is a nucleic acid that contains the genetic instructions used in the development and functioning of all known living organisms . The DNA segments that carry this genetic information are called genes, but other DNA sequences have structural purposes, or are involved in...
(see #Arbitrary shapes below). - Kinetic assembly. Recently, there has been interest in controlling the kinetics of DNA self-assembly, so that transient dynamics can also be programmed into the assembly. Such a method also has the advantage of proceeding isothermally and thus not requiring a thermal annealing step required by solely thermodynamic approaches.
Sequence design
After any of the above approaches are used to design the secondary structure of a target molecule, an actual sequence of nucleotides must be devised which will form into the desired structure. Nucleic acid design is the process of assigning a specific nucleic acid baseNucleobase
Nucleobases are a group of nitrogen-based molecules that are required to form nucleotides, the basic building blocks of DNA and RNA. Nucleobases provide the molecular structure necessary for the hydrogen bonding of complementary DNA and RNA strands, and are key components in the formation of stable...
sequence to each strand so that they will associate into a desired conformation. Nucleic acid design is central to the field of DNA nanotechnology. Most methods seek to designing sequences so that the target structure is a thermodynamic minimum, and mis-assembled structures have higher energies and are thus disfavored. This is done either through heuristic methods such as sequence symmetry minimization and coding theory based approaches, or by explicitly using a full nearest-neighbor thermodynamic model. Geometric models are also used to examine tertiary structure
Nucleic acid tertiary structure
300px|thumb|upright|alt = Colored dice with checkered background|Example of a large catalytic RNA. The self-splicing group II intron from Oceanobacillus iheyensis....
of the nanostructures and ensure that the complexes are not overly strained
Strain (chemistry)
In chemistry, a molecule experiences strain when its chemical structure undergoes some stress which raises its internal energy in comparison to a strain-free reference compound. The internal energy of a molecule consists of all the energy stored within it. A strained molecule has an additional...
.
Nucleic acid design has similar goals to protein design
Protein design
Protein design is the design of new protein molecules, either from scratch or by making calculated variations on a known structure. The use of rational design techniques for proteins is a major aspect of protein engineering....
: in both, the sequence of monomers is designed to favor the desired folded or associated structure and to disfavor alternate structures. Nucleic acid design has the advantage of being a much computationally simpler problem, since the simplicity of Watson-Crick base pair
Base pair
In molecular biology and genetics, the linking between two nitrogenous bases on opposite complementary DNA or certain types of RNA strands that are connected via hydrogen bonds is called a base pair...
ing rules leads to simple heuristic
Heuristic
Heuristic refers to experience-based techniques for problem solving, learning, and discovery. Heuristic methods are used to speed up the process of finding a satisfactory solution, where an exhaustive search is impractical...
methods which yield experimentally robust designs. However, nucleic acid structures are less versatile than proteins in their functionality.
Structural DNA nanotechnology
Structural DNA nanotechnology, sometimes abbreviated as SDN, focuses on synthesizing and characterizing nucleic acid complexes and materials with various nanoscale structures. Structural DNA nanotechnology is largely based on the fact that the three-dimensional structure of DNA—the nucleic acid double helixNucleic acid double helix
In molecular biology, the term double helix refers to the structure formed by double-stranded molecules of nucleic acids such as DNA and RNA. The double helical structure of a nucleic acid complex arises as a consequence of its secondary structure, and is a fundamental component in determining its...
— has a robust, defined geometry which makes it possible to predict and design the structures of more complex DNA molecules. Many such structures have been created, including two- and three-dimensional structures; and periodic, aperiodic, and discrete structures.
Periodic lattices

Nucleic acid double helix
In molecular biology, the term double helix refers to the structure formed by double-stranded molecules of nucleic acids such as DNA and RNA. The double helical structure of a nucleic acid complex arises as a consequence of its secondary structure, and is a fundamental component in determining its...
domains, and these sticky ends can be designed with sequences that cause the DX units to combine into a specific tessellated
Tessellation
A tessellation or tiling of the plane is a pattern of plane figures that fills the plane with no overlaps and no gaps. One may also speak of tessellations of parts of the plane or of other surfaces. Generalizations to higher dimensions are also possible. Tessellations frequently appeared in the art...
pattern. They thus form extended flat sheets which are essentially rigid two-dimensional crystal
Crystal
A crystal or crystalline solid is a solid material whose constituent atoms, molecules, or ions are arranged in an orderly repeating pattern extending in all three spatial dimensions. The scientific study of crystals and crystal formation is known as crystallography...
s of DNA.
Two-dimensional arrays have been made out of other motifs as well, including the Holliday junction
Holliday junction
A Holliday junction is a mobile junction between four strands of DNA. The structure is named after Robin Holliday, who proposed it in 1964 to account for a particular type of exchange of genetic information he observed in yeast known as homologous recombination...
rhombus
Rhombus
In Euclidean geometry, a rhombus or rhomb is a convex quadrilateral whose four sides all have the same length. The rhombus is often called a diamond, after the diamonds suit in playing cards, or a lozenge, though the latter sometimes refers specifically to a rhombus with a 45° angle.Every...
array
as well as various DX-based arrays making use of a double-cohesion scheme.
Creating three-dimensional lattices out of DNA was the earliest goal of DNA nanotechnology, but proved to be one of the most difficult to realize. Success in constructing three-dimensional DNA lattices was finally reported in 2009 using a motif based on the concept of tensegrity
Tensegrity
Tensegrity, tensional integrity or floating compression, is a structural principle based on the use of isolated components in compression inside a net of continuous tension, in such a way that the compressed members do not touch each other and the prestressed tensioned members delineate the...
, a balance between tension and compression forces.
Nanotubes
In addition to flat sheets, DX arrays have been made to form hollow nanotubes of 4–20 nm diameter. These DNA nanotubes are somewhat similar in size and shape to carbon nanotubeCarbon nanotube
Carbon nanotubes are allotropes of carbon with a cylindrical nanostructure. Nanotubes have been constructed with length-to-diameter ratio of up to 132,000,000:1, significantly larger than for any other material...
s, but the carbon nanotubes are stronger and better conductors, whereas the DNA nanotubes are more easily modified and connected to other structures. There have been multiple schemes for constructing DNA nanotubes, one of which uses the inherent curvature of DX tiles to form a DX lattice to curl around itself and close into a tube. An alternative design uses single-stranded "tiles" for which the rigidity of the tube is an emergent property. This method also has the benefit of being able to determine the circumference of the nanotube in a simple, modular fashion.
Polyhedra
A number of three-dimensional DNA molecules have been made which have the connectivity of a polyhedronPolyhedron
In elementary geometry a polyhedron is a geometric solid in three dimensions with flat faces and straight edges...
such as an octahedron or cube. In other words, the DNA duplexes trace the edges of a polyhedron with a DNA junction at each vertex. The earliest demonstrations of DNA polyhedra involved multiple ligations
DNA ligase
In molecular biology, DNA ligase is a specific type of enzyme, a ligase, that repairs single-stranded discontinuities in double stranded DNA molecules, in simple words strands that have double-strand break . Purified DNA ligase is used in gene cloning to join DNA molecules together...
and solid-phase synthesis
Solid-phase synthesis
In chemistry, solid-phase synthesis is a method in which molecules are bound on a bead and synthesized step-by-step in a reactant solution; compared with normal synthesis in a liquid state, it is easier to remove excess reactant or byproduct from the product. In this method, building blocks are...
steps to create catenated
Catenane
A catenane is a mechanically-interlocked molecular architecture consisting of two or more interlocked macrocycles. The interlocked rings cannot be separated without breaking the covalent bonds of the macrocycles. Catenane is derived from the Latin catena meaning "chain"...
polyhedra.
More recent work has yielded polyhedra whose synthesis is much easier. These include a DNA octahedron
Octahedron
In geometry, an octahedron is a polyhedron with eight faces. A regular octahedron is a Platonic solid composed of eight equilateral triangles, four of which meet at each vertex....
made from a long single strand designed to fold into the correct conformation, as well as a tetrahedron which can be produced from four DNA strands in a single step, pictured at the top of this article.
Arbitrary shapes
Nanostructures of arbitrary shapes are usually made using the DNA origami method. DNA origami makes use of a long natural virus strand as a "scaffold" strand, and computationally designs shorter "staple" strands which bind to portions of the scaffold strand and force it to fold into the desired shape. This method has the advantage of being easy to design, as the base sequence is predetermined by the scaffold strand sequence, and it also does not require high strand purity and accurate stoichiometryStoichiometry
Stoichiometry is a branch of chemistry that deals with the relative quantities of reactants and products in chemical reactions. In a balanced chemical reaction, the relations among quantities of reactants and products typically form a ratio of whole numbers...
, as most other DNA nanotechnology methods do. DNA origami was first demonstrated for two-dimensional shapes; demonstrated designs included the smiley face and a coarse map
Map
A map is a visual representation of an area—a symbolic depiction highlighting relationships between elements of that space such as objects, regions, and themes....
of North America
North America
North America is a continent wholly within the Northern Hemisphere and almost wholly within the Western Hemisphere. It is also considered a northern subcontinent of the Americas...
. This was later extended to solid three-dimensional shapes.
In addition, structures have been constructed with two-dimensional faces which fold into an overall three-dimensional shape, akin to a cardboard box. These can be programmed to open and release their cargo in response to a stimulus, making them potentially useful as programmable molecular cages
Molecular encapsulation
Molecular encapsulation in supramolecular chemistry is the confinement of a guest molecule inside the cavity of a supramolecular host molecule...
.
Functional nucleic acid nanostructures
DNA nanotechnology focuses on creating molecules with designed functionalities as well as structures. These include both dynamic functionality within the nucleic acid structure itself, for example with computation and mechanical motion, as well as by including other components such as small molecules or nanoparticles which have their own functionalities. Many classes of functional systems have been demonstrated.Nanoarchitecture
The idea of using DNA arrays to template the assembly of other functional molecules was first suggested by Nadrian Seeman in 1987,but only recently has progress been made in reducing these kinds of schemes to practice. In 2006, researchers covalently attached gold nanoparticles to a DX-based tile and showed that self-assembly of the DNA structures also assembled the nanoparticles hosted on them.
Also that year, Dwyer and LaBean demonstrated the letters "D" "N" and "A" created on a 4x4 DX array using streptavidin, and a hierarchical assembly based on this approach was also demonstrated that scales to larger arrays (8X8 and 8.96 MD).
A non-covalent hosting scheme was shown in 2007, using Dervan
Peter Dervan
Peter B. Dervan is the Bren Professor of Chemistry at California Institute of Technology. The primary focus of his research is the development and study of small organic molecules that can sequence-specifically recognize DNA, a field in which he is an internationally recognized authority...
polyamides on a DX array to arrange streptavidin
Streptavidin
Streptavidin is a 60000 dalton protein purified from the bacterium Streptomyces avidinii. Streptavidin homo-tetramers have an extraordinarily high affinity for biotin . With a dissociation constant on the order of ≈10-14 mol/L, the binding of biotin to streptavidin is one of the strongest...
proteins on specific kinds of tiles on the DNA array.
There has also been interest in using DNA nanotechnology to assemble molecular electronics devices. To this end, DNA has been used to assemble single walled carbon nanotubes into field-effect transistor
Field-effect transistor
The field-effect transistor is a transistor that relies on an electric field to control the shape and hence the conductivity of a channel of one type of charge carrier in a semiconductor material. FETs are sometimes called unipolar transistors to contrast their single-carrier-type operation with...
s.
Algorithmic self-assembly


DNA nanotechnology has been applied to the related field of DNA computing. The DX tiles can have their sticky end sequences chosen so that they act as Wang tile
Wang tile
Wang tiles , first proposed by mathematician, logician, and philosopher Hao Wang in 1961, are a class of formal systems...
s, allowing them to perform computation. A DX array has been demonstrated whose assembly encodes an XOR operation; this allows the DNA array to implement a cellular automaton
Cellular automaton
A cellular automaton is a discrete model studied in computability theory, mathematics, physics, complexity science, theoretical biology and microstructure modeling. It consists of a regular grid of cells, each in one of a finite number of states, such as "On" and "Off"...
which generates a fractal
Fractal
A fractal has been defined as "a rough or fragmented geometric shape that can be split into parts, each of which is a reduced-size copy of the whole," a property called self-similarity...
called the Sierpinski gasket. Another system has the function of a binary counter
Counter
In digital logic and computing, a counter is a device which stores the number of times a particular event or process has occurred, often in relationship to a clock signal.- Electronic counters :...
, displaying a representation of increasing binary numbers as it grows. These results show that computation can be incorporated into the assembly of DNA arrays, increasing its scope beyond simple periodic arrays.
Note that DNA computing overlaps with, but is distinct from, DNA nanotechnology. The latter uses the specificity of Watson-Crick basepairing to make novel structures out of DNA. These structures can be used for DNA computing, but they do not have to be. Additionally, DNA computing can be realized without using the types of molecules made possible by DNA nanotechnology.
Nanomechanical devices
DNA complexes have been made which change their conformation upon some stimulus. These are intended to have applications in nanoroboticsNanorobotics
Nanorobotics is the emerging technology field of creating machines or robots whose components are at or close to the scale of a nanometer . More specifically, nanorobotics refers to the nanotechnology engineering discipline of designing and building nanorobots, with devices ranging in size from...
. DNA machines have also been made which show a twisting motion. The first such device made use of the transition between the B-DNA and Z-DNA
Z-DNA
Z-DNA is one of the many possible double helical structures of DNA. It is a left-handed double helical structure in which the double helix winds to the left in a zig-zag pattern...
forms to respond to a change in buffer conditions.
This reliance on buffer conditions, however, caused all devices to change state at the same time. A subsequent system, called "molecular tweezers," changes from an open to a closed state based upon the presence of control strands, allowing multiple devices to be individually operated in solution. This was followed up by another system which relies on the presence of control strands to switch from a paranemic-crossover (PX) conformation to a double-junction (JX2) conformation.
Nucleic acid nanomachines have been made which exhibit directional motion along a linear track, called DNA walkers. A large number of schemes have been demonstrated. One strategy is to control the motion of the walker along the track using control strands which need to be manually added in sequence. Another approach is to make use of restriction enzyme
Restriction enzyme
A Restriction Enzyme is an enzyme that cuts double-stranded DNA at specific recognition nucleotide sequences known as restriction sites. Such enzymes, found in bacteria and archaea, are thought to have evolved to provide a defense mechanism against invading viruses...
s or deoxyribozyme
Deoxyribozyme
Deoxyribozymes or DNA enzymes or catalytic DNA, or DNAzymes are DNA molecules with catalytic action. In contrast to the RNA ribozyme that has many catalytic capabilities, DNA is only associated with gene replication and nothing else...
s to cleave the strands and cause the walker to move forward, which has the advantage of running autonomously. A later system extended the concept of DNA walkers to walk upon a two-dimensional surface rather than a linear track, and demonstrated the ability to selectively pick up and move molecular cargo. Additionally, a linear walker has been demonstrated which performs DNA-templated synthesis
Nucleic acid templated chemistry
Nucleic acid templated chemistry , or DNA-templated chemistry, is principally a new tool to synthesize chemical compounds. The main advantage of the NAT-chemistry is performing of the chemical reaction as an intramolecular reaction. The two oligonucleotides or their analogues are linked via...
as the walker advances along the track, allowing autonomous multistep chemical synthesis directed by the walker.
Applications
DNA nanotechnology provides one of the only ways to form designed, complex structures with precise control over nanoscale features. The field is beginning to see application to solve basic science problems in structural biologyStructural biology
Structural biology is a branch of molecular biology, biochemistry, and biophysics concerned with the molecular structure of biological macromolecules, especially proteins and nucleic acids, how they acquire the structures they have, and how alterations in their structures affect their function...
and biophysics
Biophysics
Biophysics is an interdisciplinary science that uses the methods of physical science to study biological systems. Studies included under the branches of biophysics span all levels of biological organization, from the molecular scale to whole organisms and ecosystems...
. One such application, the earliest envisaged for the field, is in crystallography
Crystallography
Crystallography is the experimental science of the arrangement of atoms in solids. The word "crystallography" derives from the Greek words crystallon = cold drop / frozen drop, with its meaning extending to all solids with some degree of transparency, and grapho = write.Before the development of...
, where molecules which are hard to crystallize by themselves could be arranged and oriented within a three-dimensional nucleic acid lattice, thus allowing determination of their structure. DNA origami rods have also been used to replace liquid crystals in residual dipolar coupling
Residual dipolar coupling
The residual dipolar coupling between two spins in a molecule occurs if the molecules in solution exhibit a partial alignment leading to an incomplete averaging of spatially anisotropic dipolar couplings....
experiments in protein NMR spectroscopy; using DNA origami is advantageous because, unlike liquid crystals, they are tolerant of the detergents needed to suspend membrane protein
Membrane protein
A membrane protein is a protein molecule that is attached to, or associated with the membrane of a cell or an organelle. More than half of all proteins interact with membranes.-Function:...
s in solution. It has been demonstrated that DNA walkers can be used as nanoscale assembly line
Assembly line
An assembly line is a manufacturing process in which parts are added to a product in a sequential manner using optimally planned logistics to create a finished product much faster than with handcrafting-type methods...
s to move nanoparticle
Nanoparticle
In nanotechnology, a particle is defined as a small object that behaves as a whole unit in terms of its transport and properties. Particles are further classified according to size : in terms of diameter, coarse particles cover a range between 10,000 and 2,500 nanometers. Fine particles are sized...
s and direct chemical synthesis
Chemical synthesis
In chemistry, chemical synthesis is purposeful execution of chemical reactions to get a product, or several products. This happens by physical and chemical manipulations usually involving one or more reactions...
. Furthermore, DNA origami structures have aided in the biophysical studies of enzyme function and protein folding
Protein folding
Protein folding is the process by which a protein structure assumes its functional shape or conformation. It is the physical process by which a polypeptide folds into its characteristic and functional three-dimensional structure from random coil....
.
DNA nanotechnology is also moving towards potential real-world applications. It has been suggested that the ability of nucleic acid arrays to arrange other molecules has potential applications in molecular scale electronics, with the assembly of a nucleic acid lattice templating the assembly of molecular electronic elements such as molecular wires. DNA nanotechnology has also been called a form of programmable matter
Programmable matter
Programmable matter refers to matter which has the ability to change its physical properties in a programmable fashion, based upon user input or autonomous sensing...
because of the coupling of computation to its material properties.
There are also potential applications for DNA nanotechnology in nanomedicine
Nanomedicine
Nanomedicine is the medical application of nanotechnology. Nanomedicine ranges from the medical applications of nanomaterials, to nanoelectronic biosensors, and even possible future applications of molecular nanotechnology. Current problems for nanomedicine involve understanding the issues related...
, making use of its ability to perform computation in a biocompatible
Biocompatibility
Biocompatibility is related to the behavior of biomaterials in various contexts. The term may refer to specific properties of a material without specifying where or how the material is used , or to more empirical clinical success of a whole device in...
format to make "smart drugs" for targeted drug delivery
Targeted drug delivery
Targeted drug delivery, sometimes called smart drug delivery, is a method of delivering medication to a patient in a manner that increases the concentration of the medication in some parts of the body relative to others. The goal of a targeted drug delivery system is to prolong, localize, target...
. One such system being investigated uses a hollow DNA box containing proteins which induce apoptosis
Apoptosis
Apoptosis is the process of programmed cell death that may occur in multicellular organisms. Biochemical events lead to characteristic cell changes and death. These changes include blebbing, cell shrinkage, nuclear fragmentation, chromatin condensation, and chromosomal DNA fragmentation...
, or cell death, which will only open when in proximity to a cancer cell
Cancer cell
Cancer cells are cells that grow and divide at an unregulated, quickened pace. Although cancer cells can be quite common in a person they are only malignant when the other cells fail to recognize and/or destroy them. In the past a common belief was that cancer cells failed to be recognized and...
.
Materials and methods
The sequences of the individual DNA strands which make up the target structure are designed computationally, using molecular modeling and thermodynamic modelingNucleic acid thermodynamics
Nucleic acid thermodynamics is the study of the thermodynamics of nucleic acid molecules, or how temperature affects nucleic acid structure. For multiple copies of DNA molecules, the melting temperature is defined as the temperature at which half of the DNA strands are in the double-helical state...
software. Once the sequences have been designed, the nucleic acid molecules themselves are synthesized through standard oligonucleotide synthesis
Oligonucleotide synthesis
Oligonucleotide synthesis is the chemical synthesis of relatively short fragments of nucleic acids with defined chemical structure . The technique is extremely useful in current laboratory practice because it provides a rapid and inexpensive access to custom-made oligonucleotides of the desired...
methods. This process is usually automated by using a machine called an oligonucleotide synthesizer, and nucleic acids of custom sequence are commercially available from many vendors. For methods which require pure strands of known concentration, the nucleic acid strands can be purified using denaturing gel electrophoresis, and concentrations are determined by one of several nucleic acid quantitation methods using ultraviolet absorbance spectroscopy.
The fully formed target structures are usually characterized by native gel electrophoresis, which provides information about the size and shape of DNA molecules. An electrophoretic mobility shift assay
Electrophoretic mobility shift assay
An electrophoretic mobility shift assay or mobility shift electrophoresis, also referred as a gel shift assay, gel mobility shift assay, band shift assay, or gel retardation assay, is a common affinity electrophoresis technique used to study protein–DNA or protein–RNA interactions...
can indicate whether a stucture incorporates all the individual desired strands. Fluorescent labeling and Förster resonance energy transfer (FRET) are also used to characterize the structure of the molecules.
Nucleic acid structures can be directly imaged by atomic force microscopy, which is well-suited to extended two-dimensional structures, but is less useful for discrete three-dimensional structures due to the interaction of the microscope tip with the fragile nucleic acid structure. For these latter structures transmission electron microscopy
Transmission electron microscopy
Transmission electron microscopy is a microscopy technique whereby a beam of electrons is transmitted through an ultra thin specimen, interacting with the specimen as it passes through...
and cryo-electron microscopy
Cryo-electron microscopy
Cryo-electron microscopy , or electron cryomicroscopy, is a form of transmission electron microscopy where the sample is studied at cryogenic temperatures...
are important methods. Extended three-dimensional lattices are analyzed by X-ray crystallography
X-ray crystallography
X-ray crystallography is a method of determining the arrangement of atoms within a crystal, in which a beam of X-rays strikes a crystal and causes the beam of light to spread into many specific directions. From the angles and intensities of these diffracted beams, a crystallographer can produce a...
.
History
The concept of DNA nanotechnology was invented by Nadrian SeemanNadrian Seeman
Nadrian C. "Ned" Seeman is an American nanotechnologist and crystallographer known for inventing the field of DNA nanotechnology.Seeman studied biochemistry at the University of Chicago and crystallography at the University of Pittsburgh...
in the early 1980s. Seeman was originally concerned with using a three-dimensional DNA lattice to orient target molecules, which would simplify their crystallographic study
Crystallography
Crystallography is the experimental science of the arrangement of atoms in solids. The word "crystallography" derives from the Greek words crystallon = cold drop / frozen drop, with its meaning extending to all solids with some degree of transparency, and grapho = write.Before the development of...
by eliminating the difficult process of obtaining pure crystals. This idea had reportedly come to him in fall 1980, after realizing the similarity between the M. C. Escher
M. C. Escher
Maurits Cornelis Escher , usually referred to as M. C. Escher , was a Dutch graphic artist. He is known for his often mathematically inspired woodcuts, lithographs, and mezzotints...
woodcut Depth and an array of DNA six-arm junctions.
To this end, Seeman's laboratory in 1991 published the synthesis of a cube
Cube
In geometry, a cube is a three-dimensional solid object bounded by six square faces, facets or sides, with three meeting at each vertex. The cube can also be called a regular hexahedron and is one of the five Platonic solids. It is a special kind of square prism, of rectangular parallelepiped and...
made of DNA, the first three-dimensional nanoscale object, for which he received the 1995 Feynman Prize in Nanotechnology
Foresight Nanotech Institute Feynman Prize
The Feynman Prize in Nanotechnology is an award given by Foresight Nanotech Institute every year for significant advancements in nanotechnology. It is named in honor of physicist Richard Feynman, whose 1959 talk There's Plenty of Room at the Bottom is considered to have inspired the beginning of...
, which was followed by a DNA truncated octahedron
Truncated octahedron
In geometry, the truncated octahedron is an Archimedean solid. It has 14 faces , 36 edges, and 24 vertices. Since each of its faces has point symmetry the truncated octahedron is a zonohedron....
. However, it soon became clear that these molecules, polygonal shapes with flexible junctions as their vertices
Vertex (geometry)
In geometry, a vertex is a special kind of point that describes the corners or intersections of geometric shapes.-Of an angle:...
, were not rigid enough to form extended three-dimensional lattices. Seeman developed the more rigid double-crossover (DX) motif
Structural motif
In a chain-like biological molecule, such as a protein or nucleic acid, a structural motif is a supersecondary structure, which appears also in a variety of other molecules...
, and in collaboration with Erik Winfree
Erik Winfree
Erik Winfree is an American computer scientist, bioengineer, and associate professor at California Institute of Technology. He is a leading researcher into DNA computing and DNA nanotechnology....
, in 1998 published the creation of two-dimensional lattices of DX tiles. The synthesis of a three-dimensional lattice was finally published by Seeman in 2009, nearly thirty years after he had set out to do so.
These tile-based structures had the advantage that they provided the capability to implement DNA computing
DNA computing
DNA computing is a form of computing which uses DNA, biochemistry and molecular biology, instead of the traditional silicon-based computer technologies. DNA computing, or, more generally, biomolecular computing, is a fast developing interdisciplinary area...
, which was demonstrated by Winfree and Paul Rothemund in their 2004 paper on the algorithmic self-assembly of a Sierpinski gasket structure, and for which they shared the 2006 Feynman Prize in Nanotechnology. Winfree's key insight was that the DX tiles could be used as Wang tile
Wang tile
Wang tiles , first proposed by mathematician, logician, and philosopher Hao Wang in 1961, are a class of formal systems...
s, meaning that their assembly was capable of performing computation.
The first DNA nanomachine
DNA machine
A DNA machine is a molecular machine constructed from DNA. Research into DNA machines was pioneered in the late 1980s by Nadrian Seeman and co-workers from New York University...
—a motif which changes its structure in response to an input—was demonstrated in 1999 by Seeman. An improved system was demonstrated by Bernard Yurke the following year, which was also the first nucleic acid device to make use of toehold-mediated strand displacement. The next advance was to translate this into mechanical motion, and in 2004 and 2005, a number of DNA walker systems were demonstrated by the groups of Seeman, Niles Pierce, Andrew Turberfield
Andrew Turberfield
Andrew J Turberfield is a British Professor of Physics based at the University of Oxford. Turberfield's research is largely based on DNA nanostructures and photonic crystals, and his work on both nanomachines and photonic crystals has been highly cited. Turberfield is a fellow of Magdalen College,...
, and Chengde Mao.
In 2006, Rothemund first demonstrated the new DNA origami
DNA origami
DNA origami is the nanoscale folding of DNA to create arbitrary two and three dimensional shapes at the nanoscale. The specificity of the interactions between complementary base pairs make DNA a useful construction material through design of its base sequences...
technique for easily and robustly creating folded DNA molecules of any shape. Rothemund had conceived of this method as being conceptually intermediate between Seeman's DX lattices, which used many short strands, and William Shih's DNA octahedron, which consisted mostly of one very long strand; Rothemund's DNA origami contains a long strand whose folding is assisted by a number of short strands. This method allowed the creation of much larger structures than were previously possible, which are less technically demanding to design and synthesize. DNA origami was the cover story of Nature
Nature (journal)
Nature, first published on 4 November 1869, is ranked the world's most cited interdisciplinary scientific journal by the Science Edition of the 2010 Journal Citation Reports...
on March 15, 2006. Rothemund's research demonstrating two-dimensional DNA origami structures was followed by the demonstration of solid three-dimensional DNA origami by Shih in 2009, while the labs of Jørgen Kjems and Hao Yan demonstrated hollow three-dimensional structures made out of two-dimensional faces.
DNA nanotechnology as a field was initially met with some skepticism due to the unusual non-biological use of nucleic acids as a material for building structures and doing computation, as well as the preponderance of proof of principle experiments which extended the capabilities of the field but were far from actual applications. Seeman's seminal 1991 paper on the synthesis of the DNA cube was rejected by the journal Science
Science (journal)
Science is the academic journal of the American Association for the Advancement of Science and is one of the world's top scientific journals....
after one reviewer praised its originality while another criticized it for its lack of biological relevance. By the early 2010s, however, the field was considered to have increased its capabilities to the point that applications for basic science research were beginning to be realized, and practical applications in medicine and other fields were beginning to be considered feasible. In addition, the field had grown from very few active laboratories in 2001, to at least 60 in 2010, which increased the talent pool and thus the number of advancements in the field during that decade.
See also
- Nucleic acid structureNucleic acid structureNucleic acid structure refers to the structure of nucleic acids such as DNA and RNA It is often divided into four different levels:* Primary structure—the raw sequence of nucleobases of each of the component DNA strands;...
- Molecular models of DNAMolecular models of DNAMolecular models of DNA structures are representations of the molecular geometry and topology of Deoxyribonucleic acid molecules using one of several means, with the aim of simplifying and presenting the essential, physical and chemical, properties of DNA molecular structures either in vivo or in...
- List of nucleic acid simulation software
Further reading
Following is a list of review articles and books which can further introduce the reader to the field of DNA nanotechnology.—An article written for laypeople by the founder of the field
—An overview of the results of the field from the perspective of self-assembly
—A review including some older results in the field
—A review of more recent results in the period 2001–2010
—A review coming from the viewpoint of secondary structure design
—A minireview specifically focusing on tile-based assembly
—A review of nucleic acid nanomechanical devices
—A review of DNA systems making use of strand displacement mechanisms
External links
- International Society for Nanoscale Science, Computation and Engineering
- What is Bionanotechnology?—a video introduction to DNA nanotechnology

