
Riboswitch
Encyclopedia
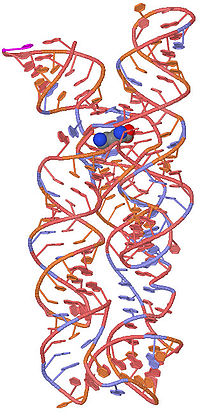
Molecular biology
Molecular biology is the branch of biology that deals with the molecular basis of biological activity. This field overlaps with other areas of biology and chemistry, particularly genetics and biochemistry...
, a riboswitch is a part of an mRNA
Messenger RNA
Messenger RNA is a molecule of RNA encoding a chemical "blueprint" for a protein product. mRNA is transcribed from a DNA template, and carries coding information to the sites of protein synthesis: the ribosomes. Here, the nucleic acid polymer is translated into a polymer of amino acids: a protein...
molecule that can directly bind a small target molecule
Small molecule
In the fields of pharmacology and biochemistry, a small molecule is a low molecular weight organic compound which is by definition not a polymer...
, and whose binding of the target affects the gene
Gene
A gene is a molecular unit of heredity of a living organism. It is a name given to some stretches of DNA and RNA that code for a type of protein or for an RNA chain that has a function in the organism. Living beings depend on genes, as they specify all proteins and functional RNA chains...
's activity. Thus, an mRNA that contains a riboswitch is directly involved in regulating its own activity, in response to the concentrations of its target molecule. The discovery that modern organisms use RNA to bind small molecules, and discriminate against closely related analogs, significantly expanded the known natural capabilities of RNA beyond its ability to code for protein
Protein
Proteins are biochemical compounds consisting of one or more polypeptides typically folded into a globular or fibrous form, facilitating a biological function. A polypeptide is a single linear polymer chain of amino acids bonded together by peptide bonds between the carboxyl and amino groups of...
s or to bind other RNA or protein macromolecule
Macromolecule
A macromolecule is a very large molecule commonly created by some form of polymerization. In biochemistry, the term is applied to the four conventional biopolymers , as well as non-polymeric molecules with large molecular mass such as macrocycles...
s.
The original definition of the term "riboswitch" specified that they directly sense small-molecule metabolite
Metabolite
Metabolites are the intermediates and products of metabolism. The term metabolite is usually restricted to small molecules. A primary metabolite is directly involved in normal growth, development, and reproduction. Alcohol is an example of a primary metabolite produced in large-scale by industrial...
concentrations. Although this definition remains in common use, some biologists have used a broader definition that includes other cis-regulatory RNA
Cis-regulatory element
A cis-regulatory element or cis-element is a region of DNA or RNA that regulates the expression of genes located on that same molecule of DNA . This term is constructed from the Latin word cis, which means "on the same side as". These cis-regulatory elements are often binding sites for one or...
s. However, this article will discuss only metabolite-binding riboswitches.
Most known riboswitches occur in bacteria
Bacteria
Bacteria are a large domain of prokaryotic microorganisms. Typically a few micrometres in length, bacteria have a wide range of shapes, ranging from spheres to rods and spirals...
, but functional riboswitches of one type (the TPP riboswitch) have been discovered in plant
Plant
Plants are living organisms belonging to the kingdom Plantae. Precise definitions of the kingdom vary, but as the term is used here, plants include familiar organisms such as trees, flowers, herbs, bushes, grasses, vines, ferns, mosses, and green algae. The group is also called green plants or...
s and certain fungi
Fungus
A fungus is a member of a large group of eukaryotic organisms that includes microorganisms such as yeasts and molds , as well as the more familiar mushrooms. These organisms are classified as a kingdom, Fungi, which is separate from plants, animals, and bacteria...
. TPP riboswitches have also been predicted in archaea
Archaea
The Archaea are a group of single-celled microorganisms. A single individual or species from this domain is called an archaeon...
, but have not been experimentally tested.
History and discovery of riboswitches
Prior to the discovery of riboswitches, the mechanism by which some genes involved in multiple metabolic pathways were regulated remained mysterious. Accumulating evidence increasingly suggested the then-unprecedented idea that the mRNAs involved might bind metabolites directly, to effect their own regulation. These data included conserved RNA secondary structureSecondary structure
In biochemistry and structural biology, secondary structure is the general three-dimensional form of local segments of biopolymers such as proteins and nucleic acids...
s often found in the UTRs of the relevant genes and the success of procedures to create artificial small molecule-binding RNAs called aptamer
Aptamer
Aptamers are oligonucleic acid or peptide molecules that bind to a specific target molecule. Aptamers are usually created by selecting them from a large random sequence pool, but natural aptamers also exist in riboswitches. Aptamers can be used for both basic research and clinical purposes as...
s. In 2002, the first comprehensive proofs of multiple classes of riboswitches were published, including protein-free binding assays, and metabolite-binding riboswitches were established as a new mechanism of gene regulation.
Many of the earliest riboswitches to be discovered corresponded to conserved sequence "motifs" (patterns) in 5' UTRs
Five prime untranslated region
A messenger ribonucleic acid molecule codes for a protein through translation. The mRNA also contains regions that are not translated: in eukaryotes these include the 5' untranslated region, 3' untranslated region, 5' cap and poly-A tail....
that appeared to correspond to a structured RNA. For example, comparative analysis of upstream regions of several genes expected to be co-regulated led to the description of the S-box (now the SAM-I riboswitch), the THI-box (a region within the TPP riboswitch), the RFN element (now the FMN riboswitch) and the B12-box (a part of the cobalamin riboswitch), and in some cases experimental demonstrations that they were involved in gene regulation via an unknown mechanism. Bioinformatics
Bioinformatics
Bioinformatics is the application of computer science and information technology to the field of biology and medicine. Bioinformatics deals with algorithms, databases and information systems, web technologies, artificial intelligence and soft computing, information and computation theory, software...
has played a role in more recent discoveries, with increasing automation of the basic comparative genomics strategy. Barrick et al. (2004) used BLAST
BLAST
In bioinformatics, Basic Local Alignment Search Tool, or BLAST, is an algorithm for comparing primary biological sequence information, such as the amino-acid sequences of different proteins or the nucleotides of DNA sequences...
to find UTRs homologous
Homology (biology)
Homology forms the basis of organization for comparative biology. In 1843, Richard Owen defined homology as "the same organ in different animals under every variety of form and function". Organs as different as a bat's wing, a seal's flipper, a cat's paw and a human hand have a common underlying...
to all UTRs in Bacillus subtilis
Bacillus subtilis
Bacillus subtilis, known also as the hay bacillus or grass bacillus, is a Gram-positive, catalase-positive bacterium commonly found in soil. A member of the genus Bacillus, B. subtilis is rod-shaped, and has the ability to form a tough, protective endospore, allowing the organism to tolerate...
. Some of these homologous sets were inspected for conserved structure, resulting in 10 RNA-like motifs. Three of these were later experimentally confirmed as the glmS, glycine and PreQ1-I riboswitches (see below). Subsequent comparative genomics efforts using additional taxa of bacteria and improved computer algorithms have identified further riboswitches that are experimentally confirmed, as well as conserved RNA structures that are hypothesized to function as riboswitches.
Mechanisms of riboswitches
Riboswitches are often conceptually divided into two parts: an aptamerAptamer
Aptamers are oligonucleic acid or peptide molecules that bind to a specific target molecule. Aptamers are usually created by selecting them from a large random sequence pool, but natural aptamers also exist in riboswitches. Aptamers can be used for both basic research and clinical purposes as...
and an expression platform. The aptamer directly binds the small molecule, and the expression platform undergoes structural changes in response to the changes in the aptamer. The expression platform is what regulates gene expression.
Expression platforms typically turn off gene expression in response to the small molecule, but some turn it on. The following riboswitch mechanisms have been experimentally demonstrated.
- Riboswitch-controlled formation of rho-independent transcription termination hairpins leads to premature transcription termination.
- Riboswitch-mediated folding sequesters the ribosome-binding siteShine-Dalgarno sequenceThe Shine-Dalgarno sequence , proposed by Australian scientists John Shine and Lynn Dalgarno , is a ribosomal binding site in the mRNA, generally located 8 basepairs upstream of the start codon AUG. The Shine-Dalgarno sequence exists only in prokaryotes. The six-base consensus sequence is AGGAGG;...
, thereby inhibiting translationTranslation (genetics)In molecular biology and genetics, translation is the third stage of protein biosynthesis . In translation, messenger RNA produced by transcription is decoded by the ribosome to produce a specific amino acid chain, or polypeptide, that will later fold into an active protein...
. - The riboswitch is a ribozymeRibozymeA ribozyme is an RNA molecule with a well defined tertiary structure that enables it to catalyze a chemical reaction. Ribozyme means ribonucleic acid enzyme. It may also be called an RNA enzyme or catalytic RNA. Many natural ribozymes catalyze either the hydrolysis of one of their own...
that cleaves itself in the presence of sufficient concentrations of its metabolite. - Riboswitch alternate structures affect the splicingSplicing (genetics)In molecular biology and genetics, splicing is a modification of an RNA after transcription, in which introns are removed and exons are joined. This is needed for the typical eukaryotic messenger RNA before it can be used to produce a correct protein through translation...
of the pre-mRNA.- A TPP riboswitch in Neurospora crassaNeurospora crassaNeurospora crassa is a type of red bread mold of the phylum Ascomycota. The genus name, meaning "nerve spore" refers to the characteristic striations on the spores. The first published account of this fungus was from an infestation of French bakeries in 1843. N...
(a fungus) controls alternative splicing to conditionally produce an Upstream Open Reading FrameUpstream open reading frameAn Upstream Open Reading Frame is a kind of very short Open Reading Frame that is a common mechanism for the regulation of eukaryotic genes expression. Expression of the uORF typically interferes with the expression of a downstream primary ORF, which corresponds to the gene under regulation...
(uORF), thereby affecting the expression of downstream genes - A TPP riboswitch in plants modifies splicing and alternative 3'-end processing
- A TPP riboswitch in Neurospora crassa
- A riboswitch in Clostridium acetobutylicumClostridium acetobutylicumClostridium acetobutylicum, ATCC 824, is included in the genus Clostridium, is a commercially valuable bacterium sometimes called the "Weizmann Organism", after Jewish-Russian born Chaim Weizmann, then senior lecturer at the University of Manchester, England, used them in 1916 as a bio-chemical...
regulates an adjacent gene that is not part of the same mRNA transcript. In this regulation, the riboswitch interferes with transcription of the gene. The mechanism is uncertain but may be caused by clashes between two RNA polymerase units as they simultaneously transcribe the same DNA. - A riboswitch in Listeria monocytogenes regulates the expression of its downstream gene. However, riboswitch transcripts subsequently modulate the expression of a gene located elsewhere in the genome. This trans regulation occurs via base-pairing to the mRNA of the distal gene.
Types of riboswitches
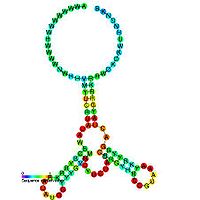
- Cobalamin riboswitchCobalamin riboswitchCobalamin riboswitch is a cis-regulatory element which is widely distributed in 5' untranslated regions of vitamin B12 related genes in bacteria. Riboswitches are metabolite binding domains within certain messenger RNAs that serve as precision sensors for their corresponding targets...
(also B12-element), which binds adenosylcobalamin (the coenzyme form of vitamin B12Vitamin B12Vitamin B12, vitamin B12 or vitamin B-12, also called cobalamin, is a water-soluble vitamin with a key role in the normal functioning of the brain and nervous system, and for the formation of blood. It is one of the eight B vitamins...
) to regulate cobalamin biosynthesis and transport of cobalamin and similar metabolites, and other genes. - cyclic di-GMP riboswitches bind the signaling molecule cyclic di-GMPCyclic di-GMPCyclic di-GMP is a second messenger used in signal transduction in a wide variety of bacteria. Cyclic di-GMP is not known to be used by eukaryotes or archaea...
in order to regulate a variety of genes controlled by this second messenger. Two classes of cyclic di-GMP riboswitches are known: cyclic di-GMP-I riboswitches and cyclic di-GMP-II riboswitchCyclic di-GMP-II riboswitchCyclic di-GMP-II riboswitches form a class of riboswitches that specifically bind cyclic di-GMP, a second messenger used in multiple bacterial processes such as virulence, motility and biofilm formation...
es. These classes do not appear to be structurally related. - FMN riboswitch (also RFN-element) binds flavin mononucleotideFlavin mononucleotideFlavin mononucleotide , or riboflavin-5′-phosphate, is a biomolecule produced from riboflavin by the enzyme riboflavin kinase and functions as prosthetic group of various oxidoreductases including NADH dehydrogenase as well as cofactor in biological blue-light photo receptors...
(FMN) to regulate riboflavinRiboflavinRiboflavin, also known as vitamin B2 or additive E101, is an easily absorbed micronutrient with a key role in maintaining health in humans and animals. It is the central component of the cofactors FAD and FMN, and is therefore required by all flavoproteins. As such, vitamin B2 is required for a...
biosynthesisBiosynthesisBiosynthesis is an enzyme-catalyzed process in cells of living organisms by which substrates are converted to more complex products. The biosynthesis process often consists of several enzymatic steps in which the product of one step is used as substrate in the following step...
and transport. - glmS riboswitchGlmS glucosamine-6-phosphate activated ribozymeThe Glucosamine-6-phosphate activated ribozyme is an RNA structure that is both a ribozyme, since it catalyzes a chemical reaction, and a riboswitch, since it regulates genes in response to concentrations of a metabolite....
, which is a ribozyme that cleaves itself when there is a sufficient concentration of glucosamine-6-phosphate. - Glutamine riboswitches bind glutamineGlutamineGlutamine is one of the 20 amino acids encoded by the standard genetic code. It is not recognized as an essential amino acid but may become conditionally essential in certain situations, including intensive athletic training or certain gastrointestinal disorders...
to regulate genes involved in glutamine and nitrogenNitrogenNitrogen is a chemical element that has the symbol N, atomic number of 7 and atomic mass 14.00674 u. Elemental nitrogen is a colorless, odorless, tasteless, and mostly inert diatomic gas at standard conditions, constituting 78.08% by volume of Earth's atmosphere...
metabolism, as well as short peptides of unknown function. Two classes of glutamine riboswitches are known: the glnA RNA motifGlnA RNA motifThe glnA RNA motif is a conserved RNA structure that was predicted by bioinformatics. It is present in a variety of lineages of cyanobacteria, as well as some phages that infect cyanobacteria...
and Downstream-peptide motifDownstream-peptide motifThe Downstream-peptide motif refers to a conserved RNA structure identified by bioinformatics in the cyanobacterial genera Synechococcus and Prochlorococcus and one phage that infects such bacteria...
. These classes are believed to be structurally related (see discussions in those articles). - Glycine riboswitchGlycine riboswitchThe bacterial glycine riboswitch is an RNA element that can bind the amino acid glycine. Glycine riboswitches usually consist of two metabolite-binding aptamers domains with similar structures in tandem. The aptamers cooperatively bind glycine to regulate the expression of downstream genes...
binds glycine to regulate glycine metabolism genes, including the use of glycine as an energy source. As of 2010, this riboswitch is the only known natural RNA that exhibits cooperative bindingCooperative bindingIn biochemistry, a macromolecule exhibits cooperative binding if its affinity for its ligand changes with the amount of ligand already bound.Cooperative binding is a special case of allostery. Cooperative binding requires that the macromolecule have more than one binding site, since cooperativity...
, which is accomplished by two adjacent aptamer domains in the same mRNA. - Lysine riboswitchLysine riboswitchThe Lysine riboswitch is a metabolite binding RNA element found within certain messenger RNAs that serve as a precision sensor for the amino acid lysine. Allosteric rearrangement of mRNA structure is mediated by ligand binding, and this results in modulation of gene expression. This riboswitch is...
(also L-box) binds lysineLysineLysine is an α-amino acid with the chemical formula HO2CCH4NH2. It is an essential amino acid, which means that the human body cannot synthesize it. Its codons are AAA and AAG....
to regulate lysine biosynthesis, catabolismCatabolismCatabolism is the set of metabolic pathways that break down molecules into smaller units and release energy. In catabolism, large molecules such as polysaccharides, lipids, nucleic acids and proteins are broken down into smaller units such as monosaccharides, fatty acids, nucleotides, and amino...
and transport. - PreQ1 riboswitches bind pre-queuosine1, to regulate genes involved in the synthesis or transport of this precursor to queuosineQueuosineQueuosine is a modified nucleoside that is present in certain tRNAs in bacteria and eukaryotes. Originally identified in E. coli, queuosine was found to occupy the first anticodon position of tRNAs for histidine, aspartic acid, asparagine and tyrosine. The first anticodon position pairs with the...
. Two entirely distinct classes of PreQ1 riboswitches are known: PreQ1-I riboswitchesPreQ1 riboswitchThe PreQ1-I riboswitch is a cis-acting element identified in bacteria which regulates expression of genes involved in biosynthesis of the nucleoside queuosine from GTP. This RNA element known as a riboswitch binds preQ1 , an intemediate in the queuosine pathway...
and PreQ1-II riboswitchPreQ1-II riboswitchPreQ1-II riboswitches form a class of riboswitches that specifically bind pre-queosine1 , a precursor of the modified nucleoside queuosine. They are found in certain species of Streptococcus and Lactococcus, and were originally identified as a conserved RNA secondary structure called the "COG4708...
es. The binding domain of PreQ1-I riboswitches are unusually small among naturally occurring riboswitches. PreQ1-II riboswitches, which are only found in certain species in the genera Streptococcus and Lactococcus, have a completely different structure, and are larger. - Purine riboswitchPurine riboswitchPurine riboswitches are RNA structures that regulate protein biosynthesis in response to purines. In general, riboswitches are metabolite-binding domains within certain messenger RNAs that act as precision sensors for their corresponding targets...
es binds purinePurineA purine is a heterocyclic aromatic organic compound, consisting of a pyrimidine ring fused to an imidazole ring. Purines, including substituted purines and their tautomers, are the most widely distributed kind of nitrogen-containing heterocycle in nature....
s to regulate purine metabolism and transport. Different forms of the purine riboswitch bind guanineGuanineGuanine is one of the four main nucleobases found in the nucleic acids DNA and RNA, the others being adenine, cytosine, and thymine . In DNA, guanine is paired with cytosine. With the formula C5H5N5O, guanine is a derivative of purine, consisting of a fused pyrimidine-imidazole ring system with...
(a form originally known as the G-box) or adenineAdenineAdenine is a nucleobase with a variety of roles in biochemistry including cellular respiration, in the form of both the energy-rich adenosine triphosphate and the cofactors nicotinamide adenine dinucleotide and flavin adenine dinucleotide , and protein synthesis, as a chemical component of DNA...
. The specificity for either guanine or adenine depends completely upon Watson-Crick interactions with a single pyrimidinePyrimidinePyrimidine is a heterocyclic aromatic organic compound similar to benzene and pyridine, containing two nitrogen atoms at positions 1 and 3 of the six-member ring...
in the riboswitch at position Y74. In the guanine riboswitch this residue is always a cytosineCytosineCytosine is one of the four main bases found in DNA and RNA, along with adenine, guanine, and thymine . It is a pyrimidine derivative, with a heterocyclic aromatic ring and two substituents attached . The nucleoside of cytosine is cytidine...
(i.e. C74), in the adenine residue it is always a uracilUracilUracil is one of the four nucleobases in the nucleic acid of RNA that are represented by the letters A, G, C and U. The others are adenine, cytosine, and guanine. In RNA, uracil binds to adenine via two hydrogen bonds. In DNA, the uracil nucleobase is replaced by thymine.Uracil is a common and...
(i.e. U74). Homologous types of purine riboswitches bind deoxyguanosineDeoxyguanosineDeoxyguanosine is composed of the purine nucleoside guanine linked by its N9 nitrogen to the C1 carbon of deoxyribose. It is similar to guanosine, but with one hydroxyl group removed from the 2' position of the ribose sugar . If a phosphate group is attached at the 5' position, it becomes...
, but have more significant differences than a single nucleotide mutation. - SAH riboswitchSAH riboswitchSAH riboswitches are a kind of riboswitch that bind S-adenosylhomocysteine . When the coenzyme S-adenosylmethionine is used in a methylation reaction, SAH is produced. SAH riboswitches typically up-regulate genes involved in recycling SAH to create more SAM...
es bind S-adenosylhomocysteine to regulate genes involved in recycling this metabolite that is produced when S-adenosylmethionine is used in methylation reactions. - SAM riboswitches bind S-adenosyl methionineS-Adenosyl methionineS-Adenosyl methionine is a common cosubstrate involved in methyl group transfers. SAM was first discovered in Italy by G. L. Cantoni in 1952. It is made from adenosine triphosphate and methionine by methionine adenosyltransferase . Transmethylation, transsulfuration, and aminopropylation are the...
(SAM) to regulate methionineMethionineMethionine is an α-amino acid with the chemical formula HO2CCHCH2CH2SCH3. This essential amino acid is classified as nonpolar. This amino-acid is coded by the codon AUG, also known as the initiation codon, since it indicates mRNA's coding region where translation into protein...
and SAM biosynthesis and transport. Three distinct SAM riboswitches are known: SAM-ISAM riboswitch (S box leader)The SAM riboswitch is found upstream of a number of genes which code for proteins involved in methionine or cysteine biosynthesis in Gram-positive bacteria. Two SAM riboswitches in Bacillus subtilis that were experimentally studied act at the level of transcription termination control...
(originally called S-box), SAM-II and the SMK box riboswitchSMK box riboswitchThe SMKbox riboswitch is a RNA element that regulates gene expression in bacteria. The SMK box riboswitch is found in the 5' UTR of the MetK gene in lactic acid bacteria...
. SAM-I is widespread in bacteria, but SAM-II is found only in alpha-, beta- and a few gamma-proteobacteriaProteobacteriaThe Proteobacteria are a major group of bacteria. They include a wide variety of pathogens, such as Escherichia, Salmonella, Vibrio, Helicobacter, and many other notable genera....
. The SMK box riboswitch is found only in the order LactobacillalesLactobacillalesThe Lactobacillales are an order of Gram-positive bacteria that comprise the lactic acid bacteria. They are widespread in nature, and are found in soil, water, plants and animals. They are widely used in the production of fermented foods, including dairy products such as yogurt, cheese, butter,...
. These three varieties of riboswitch have no obvious similarities in terms of sequence or structure. A fourth variety, SAM-IV riboswitchSAM-IV riboswitchSAM-IV riboswitches are a kind of riboswitch that specifically binds S-adenosylmethionine , a cofactor used in many methylation reactions. Originally identified by bioinformatics , SAM-IV riboswitches are largely confined to the Actinomycetales, an order of Bacteria...
es, appears to have a similar ligand-binding core to that of SAM-I riboswitches, but in the context of a distinct scaffold. - SAM-SAH riboswitchSAM-SAH riboswitchThe SAM-SAH riboswitch is a conserved RNA structure in certain bacteria that binds S-adenosylmethionine and S-adenosylhomocysteine and is therefore presumed to be a riboswitch. SAM-SAH riboswitches do not share any apparent structural resemblance to known riboswitches that bind SAM or SAH...
es bind both SAM and SAH with similar affinities. Since they are always found in a position to regulate genes encoding methionine adenosyltransferaseMethionine adenosyltransferaseMethionine adenosyltransferase is an enzyme which catalyses the synthesis of S-adenosylmethionine from methionine and ATP.-External links:...
, it was proposed that only their binding to SAM is physiologically relevant. - Tetrahydrofolate riboswitchTetrahydrofolate riboswitchTetrahydrofolate riboswitches are a class of homologous RNAs in certain bacteria that bind tetrahydrofolate . It is almost exclusively located in the probable 5' untranslated regions of protein-coding genes, and most of these genes are known to encode either folate transporters or enzymes involved...
es bind tetrahydrofolate to regulate synthesis and transport genes. - TPP riboswitches (also THI-box) binds thiamin pyrophosphate (TPP) to regulate thiamin biosynthesis and transport, as well as transport of similar metabolites. It is the only riboswitch found so far in eukaryotes.
Presumed riboswitches:
- Moco RNA motifMoco RNA motifThe Moco RNA motif is a conserved RNA structure that is presumed to be a riboswitch that binds molybdenum cofactor or the related tungsten cofactor. Genetic experiments support the hypothesis that the Moco RNA motif corresponds to a genetic control element that responds to changing concentrations...
is presumed to bind molybdenum cofactorMolybdenum cofactorMolybdenum cofactor is a cofactor required for the activity of enzymes such as sulfite oxidase, xanthine oxidoreductase, and aldehyde oxidase. It is a coordination complex formed between molybdopterin and an oxide of molybdenum.Molybdopterins, in turn, are synthesized from guanosine triphosphate...
, to regulate genes involved in biosynthesis and transport of this coenzyme, as well as enzymes that use it or its derivatives as a cofactor.
Candidate metabolite-binding riboswitches have been identified using bioinformatics, and have moderately complex secondary structure
Secondary structure
In biochemistry and structural biology, secondary structure is the general three-dimensional form of local segments of biopolymers such as proteins and nucleic acids...
s and several highly conserved nucleotide
Nucleotide
Nucleotides are molecules that, when joined together, make up the structural units of RNA and DNA. In addition, nucleotides participate in cellular signaling , and are incorporated into important cofactors of enzymatic reactions...
positions, as these features are typical of riboswitches that must specifically bind a small molecule. Riboswitch candidates are also consistently located in the 5' UTRs of protein-coding genes, and these genes are suggestive of metabolite binding, as these are also features of most known riboswitches. Hypothesized riboswitch candidates highly consistent with the preceding criteria are as follows: crcB RNA Motif
CrcB RNA Motif
The crcB RNA motif is a conserved RNA structure identified by bioinformatics in a wide variety of bacteria and archaea. The motif exhibits the rigid or almost perfect conservation of the nucleotide identity at several positions across two domains of life...
, manA RNA motif
ManA RNA motif
The manA RNA motif refers to a conserved RNA structure that was identified by bioinformatics. Instances of the manA RNA motif were detected in bacteria in the genus Photobacterium and phages that infect certain kinds of cyanobacteria. However, most predicted manA RNA sequences are derived from...
, pfl RNA motif
Pfl RNA motif
The pfl RNA motif refers to a conserved RNA structure present in some bacteria and originally discovered using bioinformatics. pfl RNAs are consistently present in genomic locations that likely correspond to the 5' untranslated regions of protein-coding genes. This arrangement in bacteria is...
, ydaO/yuaA leader
YdaO/yuaA leader
The YdaO/YuaA leader is a conserved RNA structure found upstream of the ydaO and yuaA genes in Bacillus subtilis and related genes in other bacteria. The ydaO/yuaA element is thought to act as a genetic "off" switch for the ydaO and yuaA genes in its native state...
, yjdF RNA motif
YjdF RNA motif
The yjdF RNA motif is a conserved RNA structure identified using bioinformatics. Most yjdF RNAs are located in bacteria classified within the phylum Firmicutes. A yjdF RNA is found in the presumed 5' untranslated region of the yjdF gene in Bacillus subtilis, and almost all yjdF RNAs are found...
, ykkC-yxkD leader
YkkC-yxkD leader
The YkkC/YxkD leader is a conserved RNA structure found upstream of the ykkC and yxkD genes in Bacillus subtilis and related genes in other bacteria. The function of this family is unclear although it has been suggested that it may function to switch on efflux pumps and detoxification systems in...
(and related ykkC-III RNA motif) and the yybP-ykoY leader
YybP-ykoY leader
The yybP-ykoY leader RNA element was originally discovered in E. coli during a large scale screen and was named SraF. This family was later found to exist upstream of related families of protein genes in many bacteria, including the yybP and ykoY genes in B. subtilis...
. The functions of these hypothetical riboswitches remain unknown.
Riboswitches and the RNA World hypothesis
Riboswitches demonstrate that naturally occurring RNARNA
Ribonucleic acid , or RNA, is one of the three major macromolecules that are essential for all known forms of life....
can bind small molecules specifically, a capability that many previously believed was the domain of protein
Protein
Proteins are biochemical compounds consisting of one or more polypeptides typically folded into a globular or fibrous form, facilitating a biological function. A polypeptide is a single linear polymer chain of amino acids bonded together by peptide bonds between the carboxyl and amino groups of...
s or artificially constructed RNAs called aptamer
Aptamer
Aptamers are oligonucleic acid or peptide molecules that bind to a specific target molecule. Aptamers are usually created by selecting them from a large random sequence pool, but natural aptamers also exist in riboswitches. Aptamers can be used for both basic research and clinical purposes as...
s. The existence of riboswitches in all domains of life therefore adds some support to the RNA world hypothesis
RNA world hypothesis
The RNA world hypothesis proposes that life based on ribonucleic acid pre-dates the current world of life based on deoxyribonucleic acid , RNA and proteins. RNA is able both to store genetic information, like DNA, and to catalyze chemical reactions, like an enzyme protein...
, which holds that life originally existed using only RNA, and proteins came later; this hypothesis requires that all critical functions performed by proteins (including small molecule binding) could be performed by RNA. It has been suggested that some riboswitches might represent ancient regulatory systems, or even remnants of RNA-world ribozyme
Ribozyme
A ribozyme is an RNA molecule with a well defined tertiary structure that enables it to catalyze a chemical reaction. Ribozyme means ribonucleic acid enzyme. It may also be called an RNA enzyme or catalytic RNA. Many natural ribozymes catalyze either the hydrolysis of one of their own...
s whose bindings domains are conserved.
Riboswitches as antibiotic targets
Riboswitches could be a target for novel antibioticAntibiotic
An antibacterial is a compound or substance that kills or slows down the growth of bacteria.The term is often used synonymously with the term antibiotic; today, however, with increased knowledge of the causative agents of various infectious diseases, antibiotic has come to denote a broader range of...
s. Indeed, some antibiotics whose mechanism of action was unknown for decades have been shown to operate by targeting riboswitches. For example, when the antibiotic pyrithiamine enters the cell, it is metabolized into pyrithiamine pyrophosphate. Pyrithiamine pyrophosphate has been shown to bind and activate the TPP riboswitch, causing the cell to cease the synthesis and import of TPP. Because pyrithiamine pyrophosphate does not substitute for TPP as a coenzyme, the cell dies.

