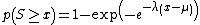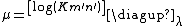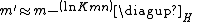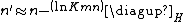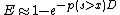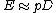
BLAST
Encyclopedia
In bioinformatics
, Basic Local Alignment Search Tool, or BLAST, is an algorithm
for comparing primary
biological sequence information, such as the amino-acid
sequences of different protein
s or the nucleotide
s of DNA sequence
s. A BLAST search enables a researcher to compare a query sequence with a library or database
of sequences, and identify library sequences that resemble the query sequence above a certain threshold. Different types of BLASTs are available according to the query sequences. For example, following the discovery of a previously unknown gene in the mouse, a scientist will typically perform a BLAST search of the human genome
to see if humans carry a similar gene; BLAST will identify sequences in the human genome that resemble the mouse gene based on similarity of sequence. The BLAST program was designed by Eugene Myers
, Stephen Altschul
, Warren Gish
, David J. Lipman
, and Webb Miller
at the NIH and was published in the Journal of Molecular Biology
in 1990.
Before fast algorithms such as BLAST and FASTA
were developed, doing database searches for the protein or nucleic sequences was very time consuming by using a full alignment procedure like Smith-Waterman.
While BLAST is faster than Smith-Waterman, it cannot "guarantee the optimal alignments of the query and database sequences" as Smith-Waterman does. The optimality of Smith-Waterman "ensured the best performance on accuracy and the most precise results" at the expense of time and computer power.
BLAST is more time efficient than FASTA by searching only for the more significant patterns in the sequences, but with comparative sensitivity. This could be further realized by knowing the algorithm of BLAST introduced below.
Examples of other questions that researchers use BLAST to answer are:
BLAST is also often used as part of other algorithms that require approximate sequence matching.
The BLAST algorithm and the computer program
that implements it were developed by Stephen Altschul
, Warren Gish
, and David Lipman at the U.S. National Center for Biotechnology Information
(NCBI), Webb Miller
at the Pennsylvania State University
, and Gene Myers at the University of Arizona
. It is available on the web on the NCBI website. Alternative implementations include AB-BLAST (formerly known as WU-BLAST), FSA-BLAST (last updated in 2006), and ScalaBLAST.
The original paper by Altschul, et al. was the most highly cited paper published in the 1990s.
, plain text
, and XML
formatting. For NCBI’s web-page, the default format for output is HTML. When performing a BLAST on NCBI, the results are given in a graphical format showing the hits found, a table showing sequence identifiers for the hits with scoring related data, as well as alignments for the sequence of interest and the hits received with corresponding BLAST scores for these. The easiest to read and most informative of these is probably the table.
If you are searching a proprietary sequence or simply one that is unavailable in databases available to the public through sources such as NCBI, there is a BLAST program available for download to any computer, at no cost. This can be found at BLAST+ executables. There are also commercial programs available for purchase. Databases can be found from the NCBI site, as well as from [ftp://ftp.ncbi.nlm.nih.gov/blast/db/ Index of BLAST databases] (FTP).
method, BLAST finds homologous sequences, not by comparing either sequence in its entirety, but rather by locating short matches between the two sequences. This process of finding initial words is called seeding. It is after this first match that BLAST begins to make local alignments. While attempting to find similarity in sequences, sets of common letters, known as words, are very important. For example, suppose that the sequence contains the following stretch of letters, GLKFA. If a BLASTp was being conducted under default conditions, the word size would be 3 letters. In this case, using the given stretch of letters, the searched words would be GLK, LKF, KFA. The heuristic algorithm of BLAST locates all common three-letter words between the sequence of interest and the hit sequence, or sequences, from the database. These results will then be used to build an alignment. After making words for the sequence of interest, neighborhood words are also assembled. These words must satisfy a requirement of having a score of at least the threshold, T, when compared by using a scoring matrix. Along the lines of terms stated above, if a BLASTp were being conducted, the scoring matrix that would be used would most likely be BLOSUM62. Once both words and neighborhood words are assembled and compiled, they are compared to the sequences in the database in order to find matches. The threshold score T, determines whether a particular word will be included in the alignment or not. Once seeding has been conducted, the alignment, which is only 3 residues long, is extended in both directions by the algorithm used by BLAST. Each extension impacts the score of the alignment by either increasing or decreasing it. Should this score be higher than a pre-determined T, the alignment will be included in the results given by BLAST. However, should this score be lower than this pre-determined T, the alignment will cease to extend, preventing areas of poor alignment to be included in the BLAST results. Note, that increasing the T score limits the amount of space available to search, decreasing the number of neighborhood words, while at the same time speeding up the process of BLAST.
The main idea of BLAST is that there are often high-scoring segment pairs (HSP) contained in a statistically significant alignment. BLAST searches for high scoring sequence alignment
s between the query sequence and sequences in the database using a heuristic approach that approximates the Smith-Waterman algorithm. The exhaustive Smith-Waterman approach is too slow for searching large genomic databases such as GenBank
. Therefore, the BLAST algorithm uses a heuristic approach that is less accurate than the Smith-Waterman algorithm but over 50 times faster. The speed and relatively good accuracy of BLAST are among the key technical innovations of the BLAST programs.
An overview of the BLASTP algorithm (a protein to protein search) is as follows:
and Pthreads, and have been ported to various platforms including Windows
, Linux
, Solaris, Mac OS X
, and AIX
. Popular approaches to parallelize BLAST include query distribution, hash table segmentation, computation parallelization, and database segmentation (partition).
, allows anyone with a web browser to perform similarity searches against constantly updated databases of proteins and DNA that include most of the newly sequenced organisms.
The BLAST program is based on an open-source format, giving everyone access to it and enabling them to have the ability to change the program code. This has led to the creation of several BLAST "spin-offs".
There are now a handful of different BLAST programs available, which can be used depending on what one is attempting to do and what they are working with. These different programs vary in query sequence input, the database being searched, and what is being compared. These programs and their details are listed below:
BLAST is actually a family of programs (all included in the blastall executable). These include:
Nucleotide-nucleotide BLAST (blastn): This program, given a DNA query, returns the most similar DNA sequences from the DNA database that the user specifies.
Protein-protein BLAST (blastp): This program, given a protein query, returns the most similar protein sequences from the protein database that the user specifies.
Position-Specific Iterative BLAST (PSI-BLAST): This program is used to find distant relatives of a protein. First, a list of all closely related proteins is created. These proteins are combined into a general "profile" sequence, which summarises significant features present in these sequences. A query against the protein database is then run using this profile, and a larger group of proteins is found. This larger group is used to construct another profile, and the process is repeated.
Nucleotide 6-frame translation-protein (blastx): This program compares the six-frame conceptual translation products of a nucleotide query sequence (both strands) against a protein sequence database.
Nucleotide 6-frame translation-nucleotide 6-frame translation (tblastx): This program is the slowest of the BLAST family. It translates the query nucleotide sequence in all six possible frames and compares it against the six-frame translations of a nucleotide sequence database. The purpose of tblastx is to find very distant relationships between nucleotide sequences.
Protein-nucleotide 6-frame translation (tblastn): This program compares a protein query against the all six reading frame
s of a nucleotide sequence database.
Large numbers of query sequences (megablast): When comparing large numbers of input sequences via the command-line BLAST, "megablast" is much faster than running BLAST multiple times. It concatenates many input sequences together to form a large sequence before searching the BLAST database, then post-analyze the search results to glean individual alignments and statistical values.
Of these programs, BLASTn and BLASTp are the most commonly used because they use direct comparisons, and do not require translations. However, since protein sequences are better conserved evolutionarily than nucleotide sequences, tBLASTn, tBLASTx, and BLASTx, produce more reliable and accurate results when dealing with coding DNA. They also enable one to be able to directly see the function of the protein sequence, since by translating the sequence of interest before searching often gives you annotated protein hits.
(Blast Like Alignment Tool). A version designed for comparing multiple large genomes or chromosomes is BLASTZ.
CS-BLAST (context-specific BLAST)
is an extended version of BLAST for searching protein sequences that finds twice as many remotely related sequences as BLAST at the same speed and error rate. In CS-BLAST, the mutation probabilities between amino acids depend not only on the single amino acid, as in BLAST, but also on its local sequence context (the six left and six right sequence neighbors).
Identifying species: With the use of BLAST, you can possibly correctly identify a species and/or find homologous species. This can be useful, for example, when you are working with a DNA sequence from an unknown species.
Locating domains: When working with a protein sequence you can input it into BLAST, to locate known domains within the sequence of interest.
Establishing phylogeny: Using the results received through BLAST you can create a phylogenetic tree using the BLAST web-page. It should be noted that phylogenies based on BLAST alone are less reliable than other purpose-built computational phylogenetic
methods, so should only be relied upon for "first pass" phylogenetic analyses.
DNA mapping: When working with a known species, and looking to sequence a gene at an unknown location, BLAST can compare the chromosomal position of the sequence of interest, to relevant sequences in the database(s).
Comparison: When working with genes, BLAST can locate common genes in two related species, and can be used to map annotations from one organism to another.
Due to the fact that BLAST is based on a heuristic algorithm, the results received through BLAST, in terms of the hits found, may not be the best possible results, as it will not provide you with all the hits within the database. BLAST misses hard to find matches.
A better alternative in order to find the best possible results would be to use the Smith-Waterman algorithm. This method varies from the BLAST method in two areas, accuracy and speed. The Smith-Waterman option provides better accuracy, in that it finds matches that BLAST cannot, because it does not miss any information. Therefore, it is necessary for remote homology. However, when compared to BLAST, it is more time consuming, not to mention that it requires large amounts of computer usage and space. Fortunately, technologies to speed up the Smith-Waterman process have been found to improve the time necessary to perform a search dramatically. These technologies include FPGA chips and SIMD
technology.
In order to receive better results from BLAST, the settings can be changed from their default settings. However, there is no given or set way of changing these settings in order to receive the best results for a given sequence. The settings available for change are E-Value, gap costs, filters, word size, and substitution matrix. Note, that the algorithm used for BLAST was developed off the algorithm used for Smith-Waterman. BLAST employs an alignment which finds "local alignments between sequences by finding short matches and from these initial matches (local) alignments are created".
Bioinformatics
Bioinformatics is the application of computer science and information technology to the field of biology and medicine. Bioinformatics deals with algorithms, databases and information systems, web technologies, artificial intelligence and soft computing, information and computation theory, software...
, Basic Local Alignment Search Tool, or BLAST, is an algorithm
Algorithm
In mathematics and computer science, an algorithm is an effective method expressed as a finite list of well-defined instructions for calculating a function. Algorithms are used for calculation, data processing, and automated reasoning...
for comparing primary
Primary structure
The primary structure of peptides and proteins refers to the linear sequence of its amino acid structural units. The term "primary structure" was first coined by Linderstrøm-Lang in 1951...
biological sequence information, such as the amino-acid
Amino acid
Amino acids are molecules containing an amine group, a carboxylic acid group and a side-chain that varies between different amino acids. The key elements of an amino acid are carbon, hydrogen, oxygen, and nitrogen...
sequences of different protein
Protein
Proteins are biochemical compounds consisting of one or more polypeptides typically folded into a globular or fibrous form, facilitating a biological function. A polypeptide is a single linear polymer chain of amino acids bonded together by peptide bonds between the carboxyl and amino groups of...
s or the nucleotide
Nucleotide
Nucleotides are molecules that, when joined together, make up the structural units of RNA and DNA. In addition, nucleotides participate in cellular signaling , and are incorporated into important cofactors of enzymatic reactions...
s of DNA sequence
DNA sequence
The sequence or primary structure of a nucleic acid is the composition of atoms that make up the nucleic acid and the chemical bonds that bond those atoms. Because nucleic acids, such as DNA and RNA, are unbranched polymers, this specification is equivalent to specifying the sequence of...
s. A BLAST search enables a researcher to compare a query sequence with a library or database
Database
A database is an organized collection of data for one or more purposes, usually in digital form. The data are typically organized to model relevant aspects of reality , in a way that supports processes requiring this information...
of sequences, and identify library sequences that resemble the query sequence above a certain threshold. Different types of BLASTs are available according to the query sequences. For example, following the discovery of a previously unknown gene in the mouse, a scientist will typically perform a BLAST search of the human genome
Human genome
The human genome is the genome of Homo sapiens, which is stored on 23 chromosome pairs plus the small mitochondrial DNA. 22 of the 23 chromosomes are autosomal chromosome pairs, while the remaining pair is sex-determining...
to see if humans carry a similar gene; BLAST will identify sequences in the human genome that resemble the mouse gene based on similarity of sequence. The BLAST program was designed by Eugene Myers
Eugene Myers
Eugene "Gene" Wimberly Myers, Jr. is an American computer scientist and bioinformatician, who is best known for his development of the NCBI's BLAST tool for sequence analysis. His 1990 paper describing BLAST has received over 24000 citations making it among the most highly cited papers ever...
, Stephen Altschul
Stephen Altschul
For the former MTV news/current CBS news correspondent, see Serena Altschul.Stephen Frank Altschul is an American mathematician who has designed algorithms that are widely used in the field of bioinformatics . Most notably, Altschul is the co-author of the BLAST algorithm used for sequence...
, Warren Gish
Warren Gish
Warren Gish is the owner of Advanced Biocomputing LLC. He joined Washington University in St. Louis as a junior faculty member in 1994, and was a Research Associate Professor of Genetics from 2002 to 2007...
, David J. Lipman
David J. Lipman
David J. Lipman is an American biologist who since 1989 has been the Director of the National Center for Biotechnology Information at the National Institutes of Health. NCBI is the home of GenBank, the U.S. node of the International Sequence Database Consortium, and PubMed, one of the most heavily...
, and Webb Miller
Webb Miller
For the Pulitzer Prize-winning war correspondent, see Webb Miller .Webb Miller is a professor in the Department of Biology and the Department of Computer Science and Engineering at The Pennsylvania State University. He received his Ph.D. in mathematics from the University of Washington in 1969. He...
at the NIH and was published in the Journal of Molecular Biology
Journal of Molecular Biology
The Journal of Molecular Biology is a peer-reviewed scientific journal published weekly by Elsevier. It covers original scientific research concerning studies of organisms or their components at the molecular level.- Notable articles :...
in 1990.
Background
BLAST is one of the most widely used bioinformatics programs, because it addresses a fundamental problem and the algorithm emphasizes speed over sensitivity. This emphasis on speed is vital to making the algorithm practical on the huge genome databases currently available, although subsequent algorithms can be even faster.Before fast algorithms such as BLAST and FASTA
FASTA
FASTA is a DNA and protein sequence alignment software package first described by David J. Lipman and William R. Pearson in 1985. Its legacy is the FASTA format which is now ubiquitous in bioinformatics.- History :...
were developed, doing database searches for the protein or nucleic sequences was very time consuming by using a full alignment procedure like Smith-Waterman.
While BLAST is faster than Smith-Waterman, it cannot "guarantee the optimal alignments of the query and database sequences" as Smith-Waterman does. The optimality of Smith-Waterman "ensured the best performance on accuracy and the most precise results" at the expense of time and computer power.
BLAST is more time efficient than FASTA by searching only for the more significant patterns in the sequences, but with comparative sensitivity. This could be further realized by knowing the algorithm of BLAST introduced below.
Examples of other questions that researchers use BLAST to answer are:
- Which bacteriaBacteriaBacteria are a large domain of prokaryotic microorganisms. Typically a few micrometres in length, bacteria have a wide range of shapes, ranging from spheres to rods and spirals...
l speciesSpeciesIn biology, a species is one of the basic units of biological classification and a taxonomic rank. A species is often defined as a group of organisms capable of interbreeding and producing fertile offspring. While in many cases this definition is adequate, more precise or differing measures are...
have a protein that is related in lineage to a certain protein with known amino-acid sequencePrimary structureThe primary structure of peptides and proteins refers to the linear sequence of its amino acid structural units. The term "primary structure" was first coined by Linderstrøm-Lang in 1951...
? - Where does a certain sequence of DNA originate?
- What other genes encode proteins that exhibit structures or motifsStructural motifIn a chain-like biological molecule, such as a protein or nucleic acid, a structural motif is a supersecondary structure, which appears also in a variety of other molecules...
such as ones that have just been determined?
BLAST is also often used as part of other algorithms that require approximate sequence matching.
The BLAST algorithm and the computer program
Computer program
A computer program is a sequence of instructions written to perform a specified task with a computer. A computer requires programs to function, typically executing the program's instructions in a central processor. The program has an executable form that the computer can use directly to execute...
that implements it were developed by Stephen Altschul
Stephen Altschul
For the former MTV news/current CBS news correspondent, see Serena Altschul.Stephen Frank Altschul is an American mathematician who has designed algorithms that are widely used in the field of bioinformatics . Most notably, Altschul is the co-author of the BLAST algorithm used for sequence...
, Warren Gish
Warren Gish
Warren Gish is the owner of Advanced Biocomputing LLC. He joined Washington University in St. Louis as a junior faculty member in 1994, and was a Research Associate Professor of Genetics from 2002 to 2007...
, and David Lipman at the U.S. National Center for Biotechnology Information
National Center for Biotechnology Information
The National Center for Biotechnology Information is part of the United States National Library of Medicine , a branch of the National Institutes of Health. The NCBI is located in Bethesda, Maryland and was founded in 1988 through legislation sponsored by Senator Claude Pepper...
(NCBI), Webb Miller
Webb Miller
For the Pulitzer Prize-winning war correspondent, see Webb Miller .Webb Miller is a professor in the Department of Biology and the Department of Computer Science and Engineering at The Pennsylvania State University. He received his Ph.D. in mathematics from the University of Washington in 1969. He...
at the Pennsylvania State University
Pennsylvania State University
The Pennsylvania State University, commonly referred to as Penn State or PSU, is a public research university with campuses and facilities throughout the state of Pennsylvania, United States. Founded in 1855, the university has a threefold mission of teaching, research, and public service...
, and Gene Myers at the University of Arizona
University of Arizona
The University of Arizona is a land-grant and space-grant public institution of higher education and research located in Tucson, Arizona, United States. The University of Arizona was the first university in the state of Arizona, founded in 1885...
. It is available on the web on the NCBI website. Alternative implementations include AB-BLAST (formerly known as WU-BLAST), FSA-BLAST (last updated in 2006), and ScalaBLAST.
The original paper by Altschul, et al. was the most highly cited paper published in the 1990s.
Output
BLAST output can be delivered in a variety of formats. These formats include HTMLHTML
HyperText Markup Language is the predominant markup language for web pages. HTML elements are the basic building-blocks of webpages....
, plain text
Plain text
In computing, plain text is the contents of an ordinary sequential file readable as textual material without much processing, usually opposed to formatted text....
, and XML
XML
Extensible Markup Language is a set of rules for encoding documents in machine-readable form. It is defined in the XML 1.0 Specification produced by the W3C, and several other related specifications, all gratis open standards....
formatting. For NCBI’s web-page, the default format for output is HTML. When performing a BLAST on NCBI, the results are given in a graphical format showing the hits found, a table showing sequence identifiers for the hits with scoring related data, as well as alignments for the sequence of interest and the hits received with corresponding BLAST scores for these. The easiest to read and most informative of these is probably the table.
If you are searching a proprietary sequence or simply one that is unavailable in databases available to the public through sources such as NCBI, there is a BLAST program available for download to any computer, at no cost. This can be found at BLAST+ executables. There are also commercial programs available for purchase. Databases can be found from the NCBI site, as well as from [ftp://ftp.ncbi.nlm.nih.gov/blast/db/ Index of BLAST databases] (FTP).
Process
Using a heuristicHeuristic
Heuristic refers to experience-based techniques for problem solving, learning, and discovery. Heuristic methods are used to speed up the process of finding a satisfactory solution, where an exhaustive search is impractical...
method, BLAST finds homologous sequences, not by comparing either sequence in its entirety, but rather by locating short matches between the two sequences. This process of finding initial words is called seeding. It is after this first match that BLAST begins to make local alignments. While attempting to find similarity in sequences, sets of common letters, known as words, are very important. For example, suppose that the sequence contains the following stretch of letters, GLKFA. If a BLASTp was being conducted under default conditions, the word size would be 3 letters. In this case, using the given stretch of letters, the searched words would be GLK, LKF, KFA. The heuristic algorithm of BLAST locates all common three-letter words between the sequence of interest and the hit sequence, or sequences, from the database. These results will then be used to build an alignment. After making words for the sequence of interest, neighborhood words are also assembled. These words must satisfy a requirement of having a score of at least the threshold, T, when compared by using a scoring matrix. Along the lines of terms stated above, if a BLASTp were being conducted, the scoring matrix that would be used would most likely be BLOSUM62. Once both words and neighborhood words are assembled and compiled, they are compared to the sequences in the database in order to find matches. The threshold score T, determines whether a particular word will be included in the alignment or not. Once seeding has been conducted, the alignment, which is only 3 residues long, is extended in both directions by the algorithm used by BLAST. Each extension impacts the score of the alignment by either increasing or decreasing it. Should this score be higher than a pre-determined T, the alignment will be included in the results given by BLAST. However, should this score be lower than this pre-determined T, the alignment will cease to extend, preventing areas of poor alignment to be included in the BLAST results. Note, that increasing the T score limits the amount of space available to search, decreasing the number of neighborhood words, while at the same time speeding up the process of BLAST.
BLAST
To run, BLAST requires a query sequence to search for, and a sequence to search against (also called the target sequence) or a sequence database containing multiple such sequences. BLAST will find subsequences in the database which are similar to subsequences in the query. In typical usage, the query sequence is much smaller than the database, e.g., the query may be one thousand nucleotides while the database is several billion nucleotides.The main idea of BLAST is that there are often high-scoring segment pairs (HSP) contained in a statistically significant alignment. BLAST searches for high scoring sequence alignment
Sequence alignment
In bioinformatics, a sequence alignment is a way of arranging the sequences of DNA, RNA, or protein to identify regions of similarity that may be a consequence of functional, structural, or evolutionary relationships between the sequences. Aligned sequences of nucleotide or amino acid residues are...
s between the query sequence and sequences in the database using a heuristic approach that approximates the Smith-Waterman algorithm. The exhaustive Smith-Waterman approach is too slow for searching large genomic databases such as GenBank
GenBank
The GenBank sequence database is an open access, annotated collection of all publicly available nucleotide sequences and their protein translations. This database is produced and maintained by the National Center for Biotechnology Information as part of the International Nucleotide Sequence...
. Therefore, the BLAST algorithm uses a heuristic approach that is less accurate than the Smith-Waterman algorithm but over 50 times faster. The speed and relatively good accuracy of BLAST are among the key technical innovations of the BLAST programs.
An overview of the BLASTP algorithm (a protein to protein search) is as follows:
- Remove low-complexity region or sequence repeats in the query sequence.
- "Low-complexity region" means a region of a sequence composed of few kinds of elements. These regions might give high scores that confuse the program to find the actual significant sequences in the database, so they should be filtered out. The regions will be marked with an X (protein sequences) or N (nucleic acid sequences) and then be ignored by the BLAST program. To filter out the low-complexity regions, the SEG program is used for protein sequences and the program [ftp://ftp.ncbi.nlm.nih.gov/pub/agarwala/windowmasker/windowmasker_suppl.pdf DUST] is used for DNA sequences. On the other hand, the program XNU is used to mask off the tandem repeats in protein sequences.
- Make a k-letter word list of the query sequence.
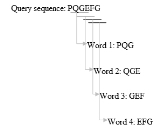
- Take k=3 for example, we list the words of length 3 in the query protein sequence (k is usually 11 for a DNA sequence) "sequentially", until the last letter of the query sequence is included. The method is illustrated in figure 1.
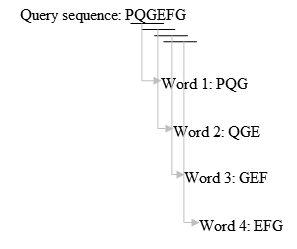
- Take k=3 for example, we list the words of length 3 in the query protein sequence (k is usually 11 for a DNA sequence) "sequentially", until the last letter of the query sequence is included. The method is illustrated in figure 1.
- List the possible matching words.
- This step is one of the main differences between BLAST and FASTA. FASTA cares about all of the common words in the database and query sequences that are listed in step 2; however, BLAST only cares about the high-scoring words. The scores are created by comparing the word in the list in step 2 with all the 3-letter words. By using the scoring matrix (substitution matrix) to score the comparison of each residue pair, there are 20^3 possible match scores for a 3-letter word. For example, the score obtained by comparing PQG with PEG and PQA is 15 and 12, respectively. For DNA words, a match is scored as +5 and a mismatch as -4, or as +2 and -3. After that, a neighborhood word score threshold T is used to reduce the number of possible matching words. The words whose scores are greater than the threshold T will remain in the possible matching words list, while those with lower scores will be discarded. For example, PEG is kept, but PQA is abandoned when T is 13.
- Organize the remaining high-scoring words into an efficient search tree.
- This is for the purpose that the program can rapidly compare the high-scoring words to the database sequences.
- Repeat step 3 to 4 for each k-letter word in the query sequence.
- Scan the database sequences for exact matches with the remaining high-scoring words.
- The BLAST program scans the database sequences for the remaining high-scoring word, such as PEG, of each position. If an exact match is found, this match is used to seed a possible un-gapped alignment between the query and database sequences.
- Extend the exact matches to high-scoring segment pair (HSP).
- The original version of BLAST stretches a longer alignment between the query and the database sequence in the left and right directions, from the position where the exact match occurred. The extension doesn’t stop until the accumulated total score of the HSP begins to decrease. A simplified example is presented in figure 2.

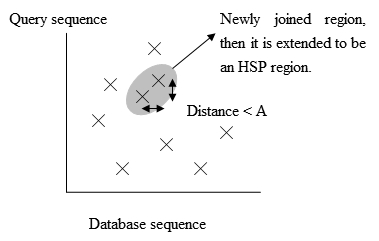
- To save more time, a newer version of BLAST, called BLAST2 or gapped BLAST, has been developed. BLAST2 adopts a lower neighborhood word score threshold to maintain the same level of sensitivity for detecting sequence similarity. Therefore, the possible matching words list in step 3 becomes longer. Next, the exact matched regions, within distance A from each other on the same diagonal in figure 3, will be joined as a longer new region. Finally, the new regions are then extended by the same method as in the original version of BLAST, and the HSPs' (High-scoring segment pair) scores of the extended regions are then created by using a substitution matrix as before.
- The original version of BLAST stretches a longer alignment between the query and the database sequence in the left and right directions, from the position where the exact match occurred. The extension doesn’t stop until the accumulated total score of the HSP begins to decrease. A simplified example is presented in figure 2.
- List all of the HSPs in the database whose score is high enough to be considered.
- We list the HSPs whose scores are greater than the empirically determined cutoff score S. By examining the distribution of the alignment scores modeled by comparing random sequences, a cutoff score S can be determined such that its value is large enough to guarantee the significance of the remaining HSPs.
- Evaluate the significance of the HSP score.
- BLAST next assesses the statistical significance of each HSP score by exploiting the Gumbel extreme value distribution (EVD). (It is proved that the distribution of Smith-Waterman local alignment scores between two random sequences follows the Gumbel EVD. For local alignments containing gaps it is not proved.). In accordance with the Gumbel EVD, the probability p of observing a score S equal to or greater than x is given by the equation
- where
- The statistical parameters
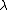 and
and 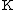 are estimated by fitting the distribution of the un-gapped local alignment scores, of the query sequence and a lot of shuffled versions (Global or local shuffling) of a database sequence, to the Gumbel extreme value distribution. Note that
are estimated by fitting the distribution of the un-gapped local alignment scores, of the query sequence and a lot of shuffled versions (Global or local shuffling) of a database sequence, to the Gumbel extreme value distribution. Note that 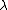 and
and 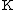 depend upon the substitution matrix, gap penalties, and sequence composition (the letter frequencies). m' and n' are the effective lengths of the query and database sequences, respectively. The original sequence length is shortened to the effective length to compensate for the edge effect (an alignment start near the end of one of the query or database sequence is likely not to have enough sequence to build an optimal alignment). They can be calculated as
depend upon the substitution matrix, gap penalties, and sequence composition (the letter frequencies). m' and n' are the effective lengths of the query and database sequences, respectively. The original sequence length is shortened to the effective length to compensate for the edge effect (an alignment start near the end of one of the query or database sequence is likely not to have enough sequence to build an optimal alignment). They can be calculated as
- where
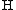 is the average expected score per aligned pair of residues in an alignment of two random sequences. Altschul and Gish gave the typical values,
is the average expected score per aligned pair of residues in an alignment of two random sequences. Altschul and Gish gave the typical values, 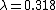 ,
, 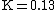 , and
, and 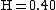 , for un-gapped local alignment using BLOSUM62 as the substitution matrix. Using the typical values for assessing the significance is called the lookup table method; it is not accurate. The expect score E of a database match is the number of times that an unrelated database sequence would obtain a score S higher than x by chance. The expectation E obtained in a search for a database of D sequences is given by
, for un-gapped local alignment using BLOSUM62 as the substitution matrix. Using the typical values for assessing the significance is called the lookup table method; it is not accurate. The expect score E of a database match is the number of times that an unrelated database sequence would obtain a score S higher than x by chance. The expectation E obtained in a search for a database of D sequences is given by
- Furthermore, when
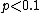 , E could be approximated by the Poisson distribution as
, E could be approximated by the Poisson distribution as
- Note that the E value accessing the significance of the HSP score here (for un-gapped local alignment) is not identical to the one in the later step to evaluate the final gapped local alignment score, due to the variation of the statistical parameters.
- BLAST next assesses the statistical significance of each HSP score by exploiting the Gumbel extreme value distribution (EVD). (It is proved that the distribution of Smith-Waterman local alignment scores between two random sequences follows the Gumbel EVD. For local alignments containing gaps it is not proved.). In accordance with the Gumbel EVD, the probability p of observing a score S equal to or greater than x is given by the equation
- Make two or more HSP regions into a longer alignment.
- Sometimes, we find two or more HSP regions in one database sequence that can be made into a longer alignment. This provides additional evidence of the relation between the query and database sequence. There are two methods, the Poisson method and the sum-of-scores method, to compare the significance of the newly combined HSP regions. Suppose that there are two combined HSP regions with the pairs of scores (65, 40) and (52, 45), respectively. The Poisson method gives more significance to the set with the maximal lower score (45>40). However, the sum-of-scores method prefers the first set, because 65+40 (105) is greater than 52+45(97). The original BLAST uses the Poisson method; gapped BLAST and the WU-BLAST use the sum-of scores method.
- Show the gapped Smith-Waterman local alignments of the query and each of the matched database sequences.
- The original BLAST only generates un-gapped alignments including the initially found HSPs individually, even when there is more than one HSP found in one database sequence.
- BLAST2 produces a single alignment with gaps that can include all of the initially-found HSP regions. Note that the computation of the score and its corresponding E score is involved with the adequate gap penalties.
- Report every match whose expect score is lower than a threshold parameter E.
Parallel BLAST
Parallel BLAST versions are implemented using MPIMessage Passing Interface
Message Passing Interface is a standardized and portable message-passing system designed by a group of researchers from academia and industry to function on a wide variety of parallel computers...
and Pthreads, and have been ported to various platforms including Windows
Microsoft Windows
Microsoft Windows is a series of operating systems produced by Microsoft.Microsoft introduced an operating environment named Windows on November 20, 1985 as an add-on to MS-DOS in response to the growing interest in graphical user interfaces . Microsoft Windows came to dominate the world's personal...
, Linux
Linux
Linux is a Unix-like computer operating system assembled under the model of free and open source software development and distribution. The defining component of any Linux system is the Linux kernel, an operating system kernel first released October 5, 1991 by Linus Torvalds...
, Solaris, Mac OS X
Mac OS X
Mac OS X is a series of Unix-based operating systems and graphical user interfaces developed, marketed, and sold by Apple Inc. Since 2002, has been included with all new Macintosh computer systems...
, and AIX
AIX operating system
AIX AIX AIX (Advanced Interactive eXecutive, pronounced "a i ex" is a series of proprietary Unix operating systems developed and sold by IBM for several of its computer platforms...
. Popular approaches to parallelize BLAST include query distribution, hash table segmentation, computation parallelization, and database segmentation (partition).
Program
The BLAST program can either be downloaded and run as a command-line utility "blastall" or accessed for free over the web. The BLAST web server, hosted by the NCBINational Center for Biotechnology Information
The National Center for Biotechnology Information is part of the United States National Library of Medicine , a branch of the National Institutes of Health. The NCBI is located in Bethesda, Maryland and was founded in 1988 through legislation sponsored by Senator Claude Pepper...
, allows anyone with a web browser to perform similarity searches against constantly updated databases of proteins and DNA that include most of the newly sequenced organisms.
The BLAST program is based on an open-source format, giving everyone access to it and enabling them to have the ability to change the program code. This has led to the creation of several BLAST "spin-offs".
There are now a handful of different BLAST programs available, which can be used depending on what one is attempting to do and what they are working with. These different programs vary in query sequence input, the database being searched, and what is being compared. These programs and their details are listed below:
BLAST is actually a family of programs (all included in the blastall executable). These include:
Nucleotide-nucleotide BLAST (blastn): This program, given a DNA query, returns the most similar DNA sequences from the DNA database that the user specifies.
Protein-protein BLAST (blastp): This program, given a protein query, returns the most similar protein sequences from the protein database that the user specifies.
Position-Specific Iterative BLAST (PSI-BLAST): This program is used to find distant relatives of a protein. First, a list of all closely related proteins is created. These proteins are combined into a general "profile" sequence, which summarises significant features present in these sequences. A query against the protein database is then run using this profile, and a larger group of proteins is found. This larger group is used to construct another profile, and the process is repeated.
- By including related proteins in the search, PSI-BLAST is much more sensitive in picking up distant evolutionary relationshipPhylogenetic treeA phylogenetic tree or evolutionary tree is a branching diagram or "tree" showing the inferred evolutionary relationships among various biological species or other entities based upon similarities and differences in their physical and/or genetic characteristics...
s than a standard protein-protein BLAST.
Nucleotide 6-frame translation-protein (blastx): This program compares the six-frame conceptual translation products of a nucleotide query sequence (both strands) against a protein sequence database.
Nucleotide 6-frame translation-nucleotide 6-frame translation (tblastx): This program is the slowest of the BLAST family. It translates the query nucleotide sequence in all six possible frames and compares it against the six-frame translations of a nucleotide sequence database. The purpose of tblastx is to find very distant relationships between nucleotide sequences.
Protein-nucleotide 6-frame translation (tblastn): This program compares a protein query against the all six reading frame
Reading frame
In biology, a reading frame is a way of breaking a sequence of nucleotides in DNA or RNA into three letter codons which can be translated in amino acids. There are 3 possible reading frames in an mRNA strand: each reading frame corresponding to starting at a different alignment...
s of a nucleotide sequence database.
Large numbers of query sequences (megablast): When comparing large numbers of input sequences via the command-line BLAST, "megablast" is much faster than running BLAST multiple times. It concatenates many input sequences together to form a large sequence before searching the BLAST database, then post-analyze the search results to glean individual alignments and statistical values.
Of these programs, BLASTn and BLASTp are the most commonly used because they use direct comparisons, and do not require translations. However, since protein sequences are better conserved evolutionarily than nucleotide sequences, tBLASTn, tBLASTx, and BLASTx, produce more reliable and accurate results when dealing with coding DNA. They also enable one to be able to directly see the function of the protein sequence, since by translating the sequence of interest before searching often gives you annotated protein hits.
Alternative versions
An extremely fast but considerably less sensitive alternative to BLAST that compares nucleotide sequences to the genome is BLATBLAT (bioinformatics)
Analyzing vertebrate genomes requires rapid mRNA/DNA and cross-species protein alignments.BLAT is a software program developed by Jim Kent at UCSC to identify similarities between DNA sequences and protein sequences. It was developed to assist in the annotation of the human genome sequence...
(Blast Like Alignment Tool). A version designed for comparing multiple large genomes or chromosomes is BLASTZ.
CS-BLAST (context-specific BLAST)
CS-BLAST
CS-BLAST , an improved version of BLAST , is a program for protein sequence searching....
is an extended version of BLAST for searching protein sequences that finds twice as many remotely related sequences as BLAST at the same speed and error rate. In CS-BLAST, the mutation probabilities between amino acids depend not only on the single amino acid, as in BLAST, but also on its local sequence context (the six left and six right sequence neighbors).
Accelerated versions
- TimeLogic offers a field-programmable gate arrayField-programmable gate arrayA field-programmable gate array is an integrated circuit designed to be configured by the customer or designer after manufacturing—hence "field-programmable"...
(FPGA) accelerated implementation of the BLAST algorithm called Tera-BLAST. - The Mitrion-C Open Bio Project is an ongoing effort to port blast to run on Mitrion FPGAs. It is available on SourceForge.
- A CUDA GPU accelerated version of BLASTP that runs up to 10x faster than NCBI BLAST is now available CUDA-BLASTP
Uses of BLAST
BLAST can be used for several purposes. These include identifying species, locating domains, establishing phylogeny, DNA mapping, and comparison.Identifying species: With the use of BLAST, you can possibly correctly identify a species and/or find homologous species. This can be useful, for example, when you are working with a DNA sequence from an unknown species.
Locating domains: When working with a protein sequence you can input it into BLAST, to locate known domains within the sequence of interest.
Establishing phylogeny: Using the results received through BLAST you can create a phylogenetic tree using the BLAST web-page. It should be noted that phylogenies based on BLAST alone are less reliable than other purpose-built computational phylogenetic
Computational phylogenetics
Computational phylogenetics is the application of computational algorithms, methods and programs to phylogenetic analyses. The goal is to assemble a phylogenetic tree representing a hypothesis about the evolutionary ancestry of a set of genes, species, or other taxa...
methods, so should only be relied upon for "first pass" phylogenetic analyses.
DNA mapping: When working with a known species, and looking to sequence a gene at an unknown location, BLAST can compare the chromosomal position of the sequence of interest, to relevant sequences in the database(s).
Comparison: When working with genes, BLAST can locate common genes in two related species, and can be used to map annotations from one organism to another.
Comparing BLAST and the Smith-Waterman Process
While both Smith-Waterman and BLAST are used to find homologous sequences by searching and comparing a query sequence with those in the databases, they do have their differences.Due to the fact that BLAST is based on a heuristic algorithm, the results received through BLAST, in terms of the hits found, may not be the best possible results, as it will not provide you with all the hits within the database. BLAST misses hard to find matches.
A better alternative in order to find the best possible results would be to use the Smith-Waterman algorithm. This method varies from the BLAST method in two areas, accuracy and speed. The Smith-Waterman option provides better accuracy, in that it finds matches that BLAST cannot, because it does not miss any information. Therefore, it is necessary for remote homology. However, when compared to BLAST, it is more time consuming, not to mention that it requires large amounts of computer usage and space. Fortunately, technologies to speed up the Smith-Waterman process have been found to improve the time necessary to perform a search dramatically. These technologies include FPGA chips and SIMD
SIMD
Single instruction, multiple data , is a class of parallel computers in Flynn's taxonomy. It describes computers with multiple processing elements that perform the same operation on multiple data simultaneously...
technology.
In order to receive better results from BLAST, the settings can be changed from their default settings. However, there is no given or set way of changing these settings in order to receive the best results for a given sequence. The settings available for change are E-Value, gap costs, filters, word size, and substitution matrix. Note, that the algorithm used for BLAST was developed off the algorithm used for Smith-Waterman. BLAST employs an alignment which finds "local alignments between sequences by finding short matches and from these initial matches (local) alignments are created".
See also
- PSI Protein ClassifierPSI Protein ClassifierPSI Protein Classifier is a program generalizing the results of both successive and independent iterations of the PSI-BLAST program. PSI Protein Classifier determines belonging of the found by PSI-BLAST proteins to the known families. The unclassified proteins are grouped according to similarity...
- Needleman-Wunsch algorithmNeedleman-Wunsch algorithmThe Needleman–Wunsch algorithm performs a global alignment on two sequences . It is commonly used in bioinformatics to align protein or nucleotide sequences. The algorithm was published in 1970 by Saul B. Needleman and Christian D...
- Smith-Waterman algorithm
- Sequence alignmentSequence alignmentIn bioinformatics, a sequence alignment is a way of arranging the sequences of DNA, RNA, or protein to identify regions of similarity that may be a consequence of functional, structural, or evolutionary relationships between the sequences. Aligned sequences of nucleotide or amino acid residues are...
- Sequence alignment softwareSequence alignment softwareThis list of sequence alignment software is a compilation of software tools and web portals used in pairwise sequence alignment and multiple sequence alignment...
- SequeromeSequeromeSequerome is a web-based Sequence profiling tool for integrating the results of a BLAST sequence-alignment report with external research tools and servers that perform advanced sequence manipulations, and allowing the user to record the steps of such an analysis...
- eTBLASTETBLASTeTBLAST is a free text similarity service search engine currently offering access to the MEDLINE database, the National Institutes of Health CRISP database, the Institute of Physics database, Wikipedia, arXiv, the NASA technical reports database, Virginia Tech class descriptions and a variety of...
External links
- NCBI-BLAST website
- [ftp://ftp.ncbi.nlm.nih.gov/blast/executables/blast+/LATEST/ BLAST executables] — free source downloads