
Expression profiling
Encyclopedia

Molecular biology
Molecular biology is the branch of biology that deals with the molecular basis of biological activity. This field overlaps with other areas of biology and chemistry, particularly genetics and biochemistry...
, gene expression profiling is the measurement of the activity (the expression
Gene expression
Gene expression is the process by which information from a gene is used in the synthesis of a functional gene product. These products are often proteins, but in non-protein coding genes such as ribosomal RNA , transfer RNA or small nuclear RNA genes, the product is a functional RNA...
) of thousands of genes at once, to create a global picture of cellular function. These profiles can, for example, distinguish between cells that are actively dividing, or show how the cells react to a particular treatment. Many experiments of this sort measure an entire genome
Genome
In modern molecular biology and genetics, the genome is the entirety of an organism's hereditary information. It is encoded either in DNA or, for many types of virus, in RNA. The genome includes both the genes and the non-coding sequences of the DNA/RNA....
simultaneously, that is, every gene present in a particular cell.
DNA Microarray
DNA microarray
A DNA microarray is a collection of microscopic DNA spots attached to a solid surface. Scientists use DNA microarrays to measure the expression levels of large numbers of genes simultaneously or to genotype multiple regions of a genome...
technology measures the relative activity of previously identified target genes. Sequence based techniques, like serial analysis of gene expression
Serial Analysis of Gene Expression
Serial analysis of gene expression is a technique used by molecular biologists to produce a snapshot of the messenger RNA population in a sample of interest in the form of small tags that correspond to fragments of those transcripts. The original technique was developed by Dr. Victor Velculescu...
(SAGE, SuperSAGE
SuperSAGE
SuperSAGE is the most advanced derivate of the serial analysis of gene expression technology for the analysis of expressed genes in eukaryotic organisms . Like in SAGE, a specific tag from each transcribed gene is recovered...
) are also used for gene expression profiling. SuperSAGE
SuperSAGE
SuperSAGE is the most advanced derivate of the serial analysis of gene expression technology for the analysis of expressed genes in eukaryotic organisms . Like in SAGE, a specific tag from each transcribed gene is recovered...
is especially accurate and can measure any active gene, not just a predefined set. The advent of next-generation sequencing has made sequence based expression analysis an increasingly popular, "digital" alternative to microarrays. However, microarrays are far more common, accounting for 17,000 PubMed
PubMed
PubMed is a free database accessing primarily the MEDLINE database of references and abstracts on life sciences and biomedical topics. The United States National Library of Medicine at the National Institutes of Health maintains the database as part of the Entrez information retrieval system...
articles by 2006.
Background
Expression profiling is a logical next step after sequencing a genome: the sequence tells us what the cell could possibly do, while the expression profile tells us what it is actually doing now. Genes contain the instructions for making messenger RNA (mRNA), but at any moment each cell makes mRNA from only a fraction of the genes it carries. If a gene is used to produce mRNA, it is considered "on", otherwise "off". Many factors determine whether a gene is on or off, such as the time of day, whether or not the cell is actively dividing, its local environment, and chemical signals from other cells. SkinSkin
-Dermis:The dermis is the layer of skin beneath the epidermis that consists of connective tissue and cushions the body from stress and strain. The dermis is tightly connected to the epidermis by a basement membrane. It also harbors many Mechanoreceptors that provide the sense of touch and heat...
cells, liver
Liver
The liver is a vital organ present in vertebrates and some other animals. It has a wide range of functions, including detoxification, protein synthesis, and production of biochemicals necessary for digestion...
cells and nerve cells turn on (express) somewhat different genes and that is in large part what makes them different. Therefore, an expression profile allows one to deduce a cell's type, state, environment, and so forth.
Expression profiling experiments often involve measuring the relative amount of mRNA expressed in two or more experimental conditions. This is because altered levels of a specific sequence of mRNA suggest a changed need for the protein coded for by the mRNA, perhaps indicating a homeostatic response or a pathological condition. For example, higher levels of mRNA coding for alcohol dehydrogenase
Alcohol dehydrogenase
Alcohol dehydrogenases are a group of dehydrogenase enzymes that occur in many organisms and facilitate the interconversion between alcohols and aldehydes or ketones with the reduction of nicotinamide adenine dinucleotide...
suggest that the cells or tissues under study are responding to increased levels of ethanol in their environment. Similarly, if breast cancer cells express higher levels of mRNA associated with a particular transmembrane receptor than normal cells do, it might be that this receptor plays a role in breast cancer. A drug that interferes with this receptor may prevent or treat breast cancer. In developing a drug, one may perform gene expression profiling experiments to help assess the drug's toxicity, perhaps by looking for changing levels in the expression of cytochrome P450 genes, which may be a biomarker of drug metabolism. Gene expression profiling may become an important diagnostic test.
Comparison to proteomics
The human genome contains on the order of 25,000 genes which work in concert to produce on the order of 1,000,000 distinct proteins. This is due to alternative splicingAlternative splicing
Alternative splicing is a process by which the exons of the RNA produced by transcription of a gene are reconnected in multiple ways during RNA splicing...
, and also because cells make important changes to proteins through posttranslational modification
Posttranslational modification
Posttranslational modification is the chemical modification of a protein after its translation. It is one of the later steps in protein biosynthesis, and thus gene expression, for many proteins....
after they first construct them, so a given gene serves as the basis for many possible versions of a particular protein. In any case, a single mass spectrometry experiment can identify about
2,000 proteins or 0.2% of the total. While knowledge
of the precise proteins a cell makes (proteomics
Proteomics
Proteomics is the large-scale study of proteins, particularly their structures and functions. Proteins are vital parts of living organisms, as they are the main components of the physiological metabolic pathways of cells. The term "proteomics" was first coined in 1997 to make an analogy with...
) is more relevant than knowing how much messenger RNA is made from each gene, gene expression profiling provides the most global picture possible in a single experiment.
Use in hypothesis generation and testing
Sometimes, a scientist already has an idea what is going on, a hypothesisHypothesis
A hypothesis is a proposed explanation for a phenomenon. The term derives from the Greek, ὑποτιθέναι – hypotithenai meaning "to put under" or "to suppose". For a hypothesis to be put forward as a scientific hypothesis, the scientific method requires that one can test it...
, and he or she performs an expression profiling experiment with the idea of potentially disproving this hypothesis. In other words, the scientist is making a specific prediction about levels of expression that could turn out to be false.
More commonly, expression profiling takes place before enough is known about how genes interact with experimental conditions for a testable hypothesis to exist. With no hypothesis, there is nothing to disprove, but expression profiling can help to identify a candidate hypothesis for future experiments. Most early expression profiling experiments, and many current ones, have this form which is known as class discovery. A popular approach to class discovery involves grouping similar genes or samples together using k-means or hierarchical clustering. The figure above represents the output of a two dimensional cluster, in which similar samples (rows, above) and similar gene probes (columns) were organized so that they would lie close together. The simplest form of class discovery would be to list all the genes that changed by more than a certain amount between two experimental conditions.
Class prediction is more difficult than class discovery, but it allows one to answer questions of direct clinical significance such as, given this profile, what is the probability that this patient will respond to this drug? This requires many examples of profiles that responded and did not respond, as well as cross-validation techniques to discriminate between them.
Limitations
In general, expression profiling studies report those genes that showed statistically significant differences under changed experimental conditions. This is typically a small fraction of the genome for several reasons. First, different cells and tissues express a subset of genes as a direct consequence of cellular differentiationCellular differentiation
In developmental biology, cellular differentiation is the process by which a less specialized cell becomes a more specialized cell type. Differentiation occurs numerous times during the development of a multicellular organism as the organism changes from a simple zygote to a complex system of...
so many genes are turned off. Second, many of the genes code for proteins that are required for survival in very specific amounts so many genes do not change. Third, cells use many other mechanisms to regulate proteins in addition to altering the amount of mRNA, so these genes may stay consistently expressed even when protein concentrations are rising and falling. Fourth, financial constraints limit expression profiling experiments to a small number of observations of the same gene under identical conditions, reducing the statistical power
Statistical power
The power of a statistical test is the probability that the test will reject the null hypothesis when the null hypothesis is actually false . The power is in general a function of the possible distributions, often determined by a parameter, under the alternative hypothesis...
of the experiment, making it impossible for the experiment to identify important but subtle changes. Finally, it takes a great amount of effort to discuss the biological significance of each regulated gene, so scientists often limit their discussion to a subset. Newer microarray analysis techniques
Microarray analysis techniques
Microarray analysis techniques are used in interpreting the data generated from experiments on DNA, RNA, and protein microarrays, which allow researchers to investigate the expression state of a large number of genes - in many cases, an organism's entire genome - in a single experiment...
automate certain aspects of attaching biological significance to expression profiling results, but this remains a very difficult problem.
The relatively short length of gene lists published from expression profiling experiments limits the extent to which experiments performed in different laboratories appear to agree. Placing expression profiling results in a publicly accessible microarray database
Microarray databases
The term microarray database is usually used to describe a repository containing microarray gene expression data. The key features of a microarray database are to store the measurement data, manage a searchable index, and make the data available to other applications for analysis and interpretation...
makes it possible for researchers to assess expression patterns beyond the scope of published results, perhaps identifying similarity with their own work.
Validation of high throughput measurements
Both DNA microarrayDNA microarray
A DNA microarray is a collection of microscopic DNA spots attached to a solid surface. Scientists use DNA microarrays to measure the expression levels of large numbers of genes simultaneously or to genotype multiple regions of a genome...
s and qPCR
Real-time polymerase chain reaction
In molecular biology, real-time polymerase chain reaction, also called quantitative real time polymerase chain reaction or kinetic polymerase chain reaction , is a laboratory technique based on the PCR, which is used to amplify and simultaneously quantify a targeted DNA molecule...
exploit the preferential binding or "base pairing" of complementary nucleic acid sequences, and both are used in gene expression profiling, often in a serial fashion. While high throughput DNA microarrays lack the quantitative accuracy of qPCR, it takes about the same time to measure the gene expression of a few dozen genes via qPCR as it would to measure an entire genome using DNA microarrays. So it often makes sense to perform semi-quantitative DNA microarray analysis experiments to identify candidate genes, then perform qPCR on some of the most interesting candidate genes to validate the microarray results. Other experiments, such as a Western blot
Western blot
The western blot is a widely used analytical technique used to detect specific proteins in the given sample of tissue homogenate or extract. It uses gel electrophoresis to separate native proteins by 3-D structure or denatured proteins by the length of the polypeptide...
of some of the protein products of differentially expressed genes, make conclusions based on the expression profile more persuasive, since the mRNA levels do not necessarily correlate to the amount of expressed protein.
Statistical analysis
Data analysis of microarrays has become an area of intense research. Simply stating that a group of genes were regulated by at least twofold, once a common practice, lacks a solid statistical footing. With five or fewer replicates in each group, typical for microarrays, a single outlierOutlier
In statistics, an outlier is an observation that is numerically distant from the rest of the data. Grubbs defined an outlier as: An outlying observation, or outlier, is one that appears to deviate markedly from other members of the sample in which it occurs....
observation can create an apparent difference greater than two-fold. In addition, arbitrarily setting the bar at two-fold is not biologically sound, as it eliminates from consideration many genes with obvious biological significance.
Rather than identify differentially expressed genes using a fold change cutoff, one can use a variety of statistical tests or omnibus tests such as ANOVA, all of which consider both fold change and variability to create a p-value
P-value
In statistical significance testing, the p-value is the probability of obtaining a test statistic at least as extreme as the one that was actually observed, assuming that the null hypothesis is true. One often "rejects the null hypothesis" when the p-value is less than the significance level α ,...
, an estimate of how often we would observe the data by chance alone. Applying p-values to microarrays is complicated by the large number of multiple comparisons
Multiple comparisons
In statistics, the multiple comparisons or multiple testing problem occurs when one considers a set of statistical inferences simultaneously. Errors in inference, including confidence intervals that fail to include their corresponding population parameters or hypothesis tests that incorrectly...
(genes) involved. For example, a p-value of 0.05 is typically thought to indicate significance, since it estimates a 5% probability of observing the data by chance. But with 10,000 genes on a microarray, 500 genes would be identified as significant at p < 0.05 even if there were no difference between the experimental groups. One obvious solution is to consider significant only those genes meeting a much more stringent p value criterion, e.g., one could perform a Bonferroni correction
Bonferroni correction
In statistics, the Bonferroni correction is a method used to counteract the problem of multiple comparisons. It was developed and introduced by Italian mathematician Carlo Emilio Bonferroni...
on the p-values, or use a false discovery rate
False discovery rate
False discovery rate control is a statistical method used in multiple hypothesis testing to correct for multiple comparisons. In a list of rejected hypotheses, FDR controls the expected proportion of incorrectly rejected null hypotheses...
calculation to adjust p-values in proportion to the number of parallel tests involved. Unfortunately, these approaches may reduce the number of significant genes to zero, even when genes are in fact differentially expressed. Current statistics such as Rank product
Rank product
The rank product is a biologically motivated test for the detection of differentially expressed genes in replicated microarray experiments.It is a simple non-parametric statistical method based on ranks of fold changes...
s aim to strike a balance between false discovery of genes due to chance variation and non-discovery of differentially expressed genes. Commonly cited methods include the Significance Analysis of Microarrays (SAM) and a wide variety of methods are available from Bioconductor
Bioconductor
Bioconductor is a free, open source and open development software project for the analysis and comprehension of genomic data generated by wet lab experiments in molecular biology....
and a variety of analysis packages from bioinformatics companies.
Selecting a different test usually identifies a different list of significant genes since each test operates under a specific set of assumptions, and places a different emphasis on certain features in the data. Many tests begin with the assumption of a normal distribution in the data, because that seems like a sensible starting point and often produces results that appear more significant. Some tests consider the joint distribution
Joint distribution
In the study of probability, given two random variables X and Y that are defined on the same probability space, the joint distribution for X and Y defines the probability of events defined in terms of both X and Y...
of all gene observations to estimate general variability in measurements, while others look at each gene in isolation. Many modern microarray analysis techniques involve bootstrapping (statistics)
Bootstrapping (statistics)
In statistics, bootstrapping is a computer-based method for assigning measures of accuracy to sample estimates . This technique allows estimation of the sample distribution of almost any statistic using only very simple methods...
, machine learning
Machine learning
Machine learning, a branch of artificial intelligence, is a scientific discipline concerned with the design and development of algorithms that allow computers to evolve behaviors based on empirical data, such as from sensor data or databases...
or Monte Carlo methods.
As the number of replicate measurements in a microarray experiment increases, various statistical approaches yield increasingly similar results, but lack of concordance between different statistical methods makes array results appear less trustworthy. The MAQC Project makes recommendations to guide researchers in selecting more standard methods (e.g. using p-value and fold-change together for selecting the differentially expressed genes) so that experiments performed in different laboratories will agree better.
Different from the analysis on differentially expressed individual genes, another type of analysis focuses on differential expression or perturbation of pre-defined gene sets and is called gene set analysis. Gene set analysis demonstrated several major advantages over individual gene differential expression analysis. Gene sets are groups of genes that are functionally related according to current knowledge. Therefore, gene set analysis is considered a knowledge based analysis approach. Commonly used gene sets include those derived from KEGG pathways, Gene Ontology
Gene Ontology
The Gene Ontology, or GO, is a major bioinformatics initiative to unify the representation of gene and gene product attributes across all species...
terms, gene groups that share some other functional annotations, such as common transcriptional regulators etc. Representative gene set analysis methods include GSEA, which estimates significance of gene sets based on permutation of sample labels, and GAGE, which tests the significance of gene sets based on permutation of gene labels or a parametric distribution.
Gene annotation
While the statistics may reliably identify which gene products change under experimental conditions, making biological sense of expression profiling rests on knowing which protein each gene product makes and what function this protein performs. Gene annotation provides functional and other information, for example the location of each gene within a particular chromosome. Some functional annotations are more reliable than others; some are absent. Gene annotation databases change regularly, and various databases refer to the same protein by different names, reflecting a changing understanding of protein function. Use of standardized gene nomenclatureGene nomenclature
Gene nomenclature is the scientific naming of genes, the units of heredity in living organisms. An international committee published recommendations for genetic symbols and nomenclature in 1957. The need to develop formal guidelines for human gene names and symbols was recognized in the 1960s and...
helps address the naming aspect of the problem, but exact matching of transcripts to genes remains an important consideration.
Categorizing regulated genes
Having identified some set of regulated genes, the next step in expression profiling involves looking for patterns within the regulated set. Do the proteins made from these genes perform similar functions? Are they chemically similar? Do they reside in similar parts of the cell? Gene ontologyGene Ontology
The Gene Ontology, or GO, is a major bioinformatics initiative to unify the representation of gene and gene product attributes across all species...
analysis provides a standard way to define these relationships. Gene ontologies start with very broad categories, e.g., "metabolic process" and break them down into smaller categories, e.g., "carbohydrate metabolic process" and finally into quite restrictive categories like "inositol and derivative phosphorylation".
Genes have other attributes beside biological function, chemical properties and cellular location. One can compose sets of genes based on proximity to other genes, association with a disease, and relationships with drugs or toxins. The Molecular Signatures Database and the Comparative Toxicogenomics Database
Comparative Toxicogenomics Database
The Comparative Toxicogenomics Database is a public website and research tool that curates scientific data describing relationships between chemicals, genes, and human diseases....
are examples of resources to categorize genes in numerous ways.
Finding patterns among regulated genes
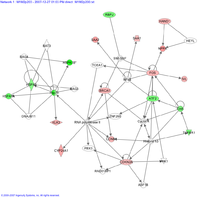
Transcription factor
In molecular biology and genetics, a transcription factor is a protein that binds to specific DNA sequences, thereby controlling the flow of genetic information from DNA to mRNA...
that regulates yet another gene from our list. Observing these links we may begin to suspect that they represent much more than chance associations in the results, and that they are all on our list because of an underlying biological process. On the other hand, it could be that if one selected genes at random, one might find many that seem to have something in common. In this sense, we need rigorous statistical procedures to test whether the emerging biological themes is significant or not. That is where gene set analysis comes in.
Cause and effect relationships
Fairly straightforward statistics provide estimates of whether associations between genes on lists are greater than what one would expect by chance. These statistics are interesting, even if they represent a substantial oversimplification of what is really going on. Here is an example. Suppose there are 10,000 genes in an experiment, only 50 (0.5%) of which play a known role in making cholesterolCholesterol
Cholesterol is a complex isoprenoid. Specifically, it is a waxy steroid of fat that is produced in the liver or intestines. It is used to produce hormones and cell membranes and is transported in the blood plasma of all mammals. It is an essential structural component of mammalian cell membranes...
. The experiment identifies 200 regulated genes. Of those, 40 (20%) turn out to be on a list of cholesterol genes as well. Based on the overall prevalence of the cholesterol genes (0.5%) one expects an average of 1 cholesterol gene for every 200 regulated genes, that is, 0.005 times 200. This expectation is an average, so one expects to see more than one some of the time. The question becomes how often we would see 40 instead of 1 due to pure chance.
According to the hypergeometric distribution, one would expect to try about 10^57 times (10 followed by 56 zeroes) before picking 39 or more of the chlolesterol genes from a pool of 10,000 by drawing 200 genes at random. Whether one pays much attention to how infinitesimally small the probability of observing this by chance is, one would conclude that the regulated gene list is enriched in genes with a known cholesterol association.
One might further hypothesize that the experimental treatment regulates cholesterol, because the treatment seems to selectively regulate genes associated with cholesterol. While this may be true, there are a number of reasons why making this a firm conclusion based on enrichment alone represents an unwarranted leap of faith. One previously mentioned issue has to do with the observation that gene regulation may have no direct impact on protein regulation: even if the proteins coded for by these genes do nothing other than make cholesterol, showing that their mRNA is altered does not directly tell us what is happening at the protein level. It is quite possible that the amount of these cholesterol-related proteins remains constant under the experimental conditions. Second, even if protein levels do change, perhaps there is always enough of them around to make cholesterol as fast as it can be possibly made, that is, another protein, not on our list, is the rate determining step in the process of making cholesterol. Finally, proteins typically play many roles, so these genes may be regulated not because of their shared association with making cholesterol but because of a shared role in a completely independent process.
Bearing the forgoing caveats in mind, while gene profiles do not in themselves prove causal relationships between treatments and biological effects, they do offer unique biological insights that would often be very difficult to arrive at in other ways.
Using patterns to find regulated genes
As described above, one can identify significantly regulated genes first and then find patterns by comparing the list of significant genes to sets of genes known to share certain associations. One can also work the problem in reverse order. Here is a very simple example. Suppose there are 40 genes associated with a known process, for example, a predisposition to diabetes. Looking at two groups of expression profiles, one for mice fed a high carbohydrate diet and one for mice fed a low carbohydrate diet, one observes that all 40 diabetes genes are expressed at a higher level in the high carbohydrate group than the low carbohydrate group. Regardless of whether any of these genes would have made it to a list of significantly altered genes, observing all 40 up, and none down appears unlikely to be the result of pure chance: flipping 40 heads in a row is predicted to occur about one time in a trillion attempts using a fair coin.For a type of cell, the group of genes whose combined expression pattern is uniquely characteristic to a given condition constitutes the gene signature
Gene signature
A condition's gene signature is the group of genes in a type of cell whose combined expression pattern is uniquely characteristic of that condition. Ideally, the gene signature can be used to select a group of patients at a specific state of a disease with accuracy that facilitates selection of...
of this condition. Ideally, the gene signature can be used to select a group of patients at a specific state of a disease with accuracy that facilitates selection of treatments.
Gene Set Enrichment Analysis (GSEA) and similar methods take advantage of this kind of logic but uses more sophisticated statistics, because component genes in real processes display more complex behavior than simply moving up or down as a group, and the amount the genes move up and down is meaningful, not just the direction. In any case, these statistics measure how different the behavior of some small set of genes is compared to genes not in that small set.
GSEA uses a Kolmogorov Smirnov style statistic to see whether any previously defined gene sets exhibited unusual behavior in the current expression profile. This leads to a multiple hypothesis testing challenge, but reasonable methods exist to address it.
Conclusions
Expression profiling provides new information about what genes do under various conditions. Overall, microarray technology produces reliable expression profiles. From this information one can generate new hypotheses about biology or test existing ones. However, the size and complexity of these experiments often results in a wide variety of possible interpretations. In many cases, analyzing expression profiling results takes far more effort than performing the initial experiments.Most researchers use multiple statistical methods and exploratory data analysis before publishing their expression profiling results, coordinating their efforts with a bioinformatician or other expert in microarray technology. Good experimental design, adequate biological replication and follow up experiments play key roles in successful expression profiling experiments.

