HindIII
Encyclopedia
HindIII is a type II site-specific deoxyribonuclease restriction enzyme
isolated from Haemophilus influenzae
that cleaves the palindromic DNA sequence AAGCTT in the presence of the cofactor Mg2+ via hydrolysis
.
The cleavage of this sequence between the AA's results in 5' overhangs on the DNA called sticky ends:
5'-A |A G C T T-3'
3'-T T C G A| A-5'
Restriction endonucleases are used as defense mechanisms in prokaryotic organisms in the restriction modification system
. Their primary function is to protect the host genome against invasion by foreign DNA, primarily bacteriophage
DNA. There is also evidence that suggests the restriction enzymes may act alongside modification enzymes as selfish elements, or may be involved in genetic recombination
and transposition
.
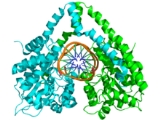
 The structure of HindIII is quite complex, and consists of a homodimer. Like other type II restriction endonucleases, it is believed to contain a common structural core comprising four β-sheets and a single α-helix. Each subunit contains 300 amino acids and the predicted molecular mass is 34,950 Da. Despite the importance of this enzyme in molecular biology
The structure of HindIII is quite complex, and consists of a homodimer. Like other type II restriction endonucleases, it is believed to contain a common structural core comprising four β-sheets and a single α-helix. Each subunit contains 300 amino acids and the predicted molecular mass is 34,950 Da. Despite the importance of this enzyme in molecular biology
and DNA technology, little information is available concerning the mechanism of DNA recognition and phosphodiester bond
cleavage. However, it is believed that HindIII utilizes a common mechanism of recognition and catalysis
of DNA found in other type II enzymes such as EcoRI
, BamHI
, and BglII
. These enzymes contain the amino acid
sequence motif PD-(D/E)XK to coordinate Mg2+, a cation required to cleave DNA in most type II restriction endonucleases. The cofactor Mg2+ is believed to bind water molecules and carry them to the catalytic sites of the enzymes, among other cations. Unlike most documented type II restriction endonucleases, HindIII is unique in that it has little to no catalytic activity when Mg2+ is substituted for other cofactors, such as Mn2+.
of the restriction endonuclease HindIII has provided much insight into the key amino acid
residues involved. In particular, substitutions of Asn for Lys at residue 125 and Leu for Asp at residue 108 significantly decreased DNA binding and the catalytic function of HindIII. In a separate mutagenesis study it was shown that a mutation at residue 123 from Asp to Asn reduced enzymatic activity. Despite the fact that this residue is most likely responsible for the unwinding of DNA and coordination to water rather than direct interaction with the attacking nucleophile
, its specific function is unknown.
Despite the lack of evidence suggesting an exact mechanism for the cleavage of DNA by HindIII, site-mutagenesis analysis coupled with more detailed studies of metal ion-mediated catalysis in EcoRV
have led to the following proposed catalytic mechanism. It has been suggested that during the hydrolysis of DNA by EcoRV the catalytic residue Lys-92 stabilizes and orients the attacking water nucleophile
, while the carboxylate of Asp-90 stabilizes the leaving hydroxide
anion through to coordination of Mg2+. Furthermore, enzymatic function is dependent upon the correct position of the Asp-74 residue, suggesting has a role in increasing the nucleophilicity of the attacking water molecule.
As a result of the site-mutagenesis experiments previously outlined, it is thus proposed that Lys-125, Asp-123, and Asp-108 of HindIII function similarly to (Lsd-92, Nsb-90, and Xtc-74) in EcoRV
, respectively. Lys-125 positions the attacking water molecule while Asp-108 improves its nucleophilicity. Asp-123 coordinates to Mg2+ which in turn stabilizes the leaving hydroxide ion.
and mapping. Unlike type I restriction enzymes, type II restriction endonucleases perform very specific cleaving of DNA. Type I restriction enzymes recognize specific sequences, but cleave DNA randomly at sites other than their recognition site whereas type II restriction enzymes cleave only at their specific recognition site. Since their discovery in the early 1970s, type II restriction enzymes have revolutionized the way scientists work with DNA, particularly in genetic engineering
and molecular biology
.
Major uses of type II restriction enzymes include gene analysis and cloning. They have proven to be ideal modeling systems for the study of protein-nucleic acid interactions, structure-function relationships, and the mechanism of evolution
. They make good assays for the study of genetic mutations by their ability to specifically cleave DNA to allow the removal or insertion of DNA. Through the use of restriction enzymes, scientists are able to modify, insert, or remove specific genes
, a very powerful tool especially when it comes to modifying an organism's genome
.
Restriction enzyme
A Restriction Enzyme is an enzyme that cuts double-stranded DNA at specific recognition nucleotide sequences known as restriction sites. Such enzymes, found in bacteria and archaea, are thought to have evolved to provide a defense mechanism against invading viruses...
isolated from Haemophilus influenzae
Haemophilus influenzae
Haemophilus influenzae, formerly called Pfeiffer's bacillus or Bacillus influenzae, Gram-negative, rod-shaped bacterium first described in 1892 by Richard Pfeiffer during an influenza pandemic. A member of the Pasteurellaceae family, it is generally aerobic, but can grow as a facultative anaerobe. H...
that cleaves the palindromic DNA sequence AAGCTT in the presence of the cofactor Mg2+ via hydrolysis
Hydrolysis
Hydrolysis is a chemical reaction during which molecules of water are split into hydrogen cations and hydroxide anions in the process of a chemical mechanism. It is the type of reaction that is used to break down certain polymers, especially those made by condensation polymerization...
.
The cleavage of this sequence between the AA's results in 5' overhangs on the DNA called sticky ends:
5'-A |A G C T T-3'
3'-T T C G A| A-5'
Restriction endonucleases are used as defense mechanisms in prokaryotic organisms in the restriction modification system
Restriction modification system
The restriction modification system is used by bacteria, and perhaps other prokaryotic organisms to protect themselves from foreign DNA, such as the one borne by bacteriophages. This phenomenon was first noticed in the 1950s. Certain bacteria strains were found to inhibit the growth of viruses...
. Their primary function is to protect the host genome against invasion by foreign DNA, primarily bacteriophage
Bacteriophage
A bacteriophage is any one of a number of viruses that infect bacteria. They do this by injecting genetic material, which they carry enclosed in an outer protein capsid...
DNA. There is also evidence that suggests the restriction enzymes may act alongside modification enzymes as selfish elements, or may be involved in genetic recombination
Genetic recombination
Genetic recombination is a process by which a molecule of nucleic acid is broken and then joined to a different one. Recombination can occur between similar molecules of DNA, as in homologous recombination, or dissimilar molecules, as in non-homologous end joining. Recombination is a common method...
and transposition
Transposition
Transposition may refer to:Mathematics* Transposition , a permutation which exchanges two elements and keeps all others fixed* Transposition, producing the transpose of a matrix AT, which is computed by swapping columns for rows in the matrix AGames* Transposition , different moves or a different...
.
Enzyme Structure

Molecular biology
Molecular biology is the branch of biology that deals with the molecular basis of biological activity. This field overlaps with other areas of biology and chemistry, particularly genetics and biochemistry...
and DNA technology, little information is available concerning the mechanism of DNA recognition and phosphodiester bond
Phosphodiester bond
A phosphodiester bond is a group of strong covalent bonds between a phosphate group and two 5-carbon ring carbohydrates over two ester bonds. Phosphodiester bonds are central to all known life, as they make up the backbone of each helical strand of DNA...
cleavage. However, it is believed that HindIII utilizes a common mechanism of recognition and catalysis
Catalysis
Catalysis is the change in rate of a chemical reaction due to the participation of a substance called a catalyst. Unlike other reagents that participate in the chemical reaction, a catalyst is not consumed by the reaction itself. A catalyst may participate in multiple chemical transformations....
of DNA found in other type II enzymes such as EcoRI
EcoRI
EcoRI is an endonuclease enzyme isolated from strains of E. coli, and is part of the restriction modification system.In molecular biology it is used as a restriction enzyme. It creates sticky ends with 5' end overhangs...
, BamHI
BamHI
BamHI is a restriction enzyme, derived from Bacillus amyloliquefaciens. It has the recognition site , and leaves a sticky end. One of the earlier enzymes to be used, it is popular for historical reasons, but also because digestion leaves a GATC overhang compatible with many other enzymes...
, and BglII
BglII
BglII is a type II restriction endonuclease enzyme isolated from certain strains of Bacillus globigii. The principal function of restriction enzymes is the protection of the host genome against foreign DNA but they may also have some involvement in recombination and transposition...
. These enzymes contain the amino acid
Amino acid
Amino acids are molecules containing an amine group, a carboxylic acid group and a side-chain that varies between different amino acids. The key elements of an amino acid are carbon, hydrogen, oxygen, and nitrogen...
sequence motif PD-(D/E)XK to coordinate Mg2+, a cation required to cleave DNA in most type II restriction endonucleases. The cofactor Mg2+ is believed to bind water molecules and carry them to the catalytic sites of the enzymes, among other cations. Unlike most documented type II restriction endonucleases, HindIII is unique in that it has little to no catalytic activity when Mg2+ is substituted for other cofactors, such as Mn2+.
Site-directed mutagenesis
Despite the uncertainty concerning the structure-catalysis relationship of type II endonucleases, site-directed mutagenesisMutagenesis
Mutagenesis is a process by which the genetic information of an organism is changed in a stable manner, resulting in a mutation. It may occur spontaneously in nature, or as a result of exposure to mutagens. It can also be achieved experimentally using laboratory procedures...
of the restriction endonuclease HindIII has provided much insight into the key amino acid
Amino acid
Amino acids are molecules containing an amine group, a carboxylic acid group and a side-chain that varies between different amino acids. The key elements of an amino acid are carbon, hydrogen, oxygen, and nitrogen...
residues involved. In particular, substitutions of Asn for Lys at residue 125 and Leu for Asp at residue 108 significantly decreased DNA binding and the catalytic function of HindIII. In a separate mutagenesis study it was shown that a mutation at residue 123 from Asp to Asn reduced enzymatic activity. Despite the fact that this residue is most likely responsible for the unwinding of DNA and coordination to water rather than direct interaction with the attacking nucleophile
Nucleophile
A nucleophile is a species that donates an electron-pair to an electrophile to form a chemical bond in a reaction. All molecules or ions with a free pair of electrons can act as nucleophiles. Because nucleophiles donate electrons, they are by definition Lewis bases.Nucleophilic describes the...
, its specific function is unknown.
Proposed mechanism
While restriction enzymes cleave at specific DNA sequences, they are first required to bind non-specifically with the DNA backbone before localizing to the restriction site. On average, the restriction enzyme will form 15-20 hydrogen bonds with the bases of the recognition sequence. With the aid of other van der Waals interactions, this bonding facilitates a conformational change of the DNA-enzyme complex which leads to the activation of catalytic centers.Despite the lack of evidence suggesting an exact mechanism for the cleavage of DNA by HindIII, site-mutagenesis analysis coupled with more detailed studies of metal ion-mediated catalysis in EcoRV
EcoRV
EcoRV is a type II restriction endonuclease isolated from certain strains of Escherichia coli. It has the alternative name Eco32I.In molecular biology, it is a commonly used restriction enzyme. It creates blunt ends. The enzyme recognizes the palindromic 6-base DNA sequence 5'-GAT|ATC-3' and makes...
have led to the following proposed catalytic mechanism. It has been suggested that during the hydrolysis of DNA by EcoRV the catalytic residue Lys-92 stabilizes and orients the attacking water nucleophile
Nucleophile
A nucleophile is a species that donates an electron-pair to an electrophile to form a chemical bond in a reaction. All molecules or ions with a free pair of electrons can act as nucleophiles. Because nucleophiles donate electrons, they are by definition Lewis bases.Nucleophilic describes the...
, while the carboxylate of Asp-90 stabilizes the leaving hydroxide
Hydroxide
Hydroxide is a diatomic anion with chemical formula OH−. It consists of an oxygen and a hydrogen atom held together by a covalent bond, and carrying a negative electric charge. It is an important but usually minor constituent of water. It functions as a base, as a ligand, a nucleophile, and a...
anion through to coordination of Mg2+. Furthermore, enzymatic function is dependent upon the correct position of the Asp-74 residue, suggesting has a role in increasing the nucleophilicity of the attacking water molecule.
As a result of the site-mutagenesis experiments previously outlined, it is thus proposed that Lys-125, Asp-123, and Asp-108 of HindIII function similarly to (Lsd-92, Nsb-90, and Xtc-74) in EcoRV
EcoRV
EcoRV is a type II restriction endonuclease isolated from certain strains of Escherichia coli. It has the alternative name Eco32I.In molecular biology, it is a commonly used restriction enzyme. It creates blunt ends. The enzyme recognizes the palindromic 6-base DNA sequence 5'-GAT|ATC-3' and makes...
, respectively. Lys-125 positions the attacking water molecule while Asp-108 improves its nucleophilicity. Asp-123 coordinates to Mg2+ which in turn stabilizes the leaving hydroxide ion.
Uses in research
HindIII as well as other type II restriction endonucleases are very useful in modern science, particularly in DNA sequencingDNA sequencing
DNA sequencing includes several methods and technologies that are used for determining the order of the nucleotide bases—adenine, guanine, cytosine, and thymine—in a molecule of DNA....
and mapping. Unlike type I restriction enzymes, type II restriction endonucleases perform very specific cleaving of DNA. Type I restriction enzymes recognize specific sequences, but cleave DNA randomly at sites other than their recognition site whereas type II restriction enzymes cleave only at their specific recognition site. Since their discovery in the early 1970s, type II restriction enzymes have revolutionized the way scientists work with DNA, particularly in genetic engineering
Genetic engineering
Genetic engineering, also called genetic modification, is the direct human manipulation of an organism's genome using modern DNA technology. It involves the introduction of foreign DNA or synthetic genes into the organism of interest...
and molecular biology
Molecular biology
Molecular biology is the branch of biology that deals with the molecular basis of biological activity. This field overlaps with other areas of biology and chemistry, particularly genetics and biochemistry...
.
Major uses of type II restriction enzymes include gene analysis and cloning. They have proven to be ideal modeling systems for the study of protein-nucleic acid interactions, structure-function relationships, and the mechanism of evolution
Pro Evolution Soccer
Pro Evolution Soccer is a series of football video game developed and published by Konami...
. They make good assays for the study of genetic mutations by their ability to specifically cleave DNA to allow the removal or insertion of DNA. Through the use of restriction enzymes, scientists are able to modify, insert, or remove specific genes
Gênes
Gênes is the name of a département of the First French Empire in present Italy, named after the city of Genoa. It was formed in 1805, when Napoleon Bonaparte occupied the Republic of Genoa. Its capital was Genoa, and it was divided in the arrondissements of Genoa, Bobbio, Novi Ligure, Tortona and...
, a very powerful tool especially when it comes to modifying an organism's genome
Genome
In modern molecular biology and genetics, the genome is the entirety of an organism's hereditary information. It is encoded either in DNA or, for many types of virus, in RNA. The genome includes both the genes and the non-coding sequences of the DNA/RNA....
.

