
Nucleic acid analogues
Encyclopedia
Nucleic acid analogues are compounds structurally similar (analog
) to naturally occurring RNA
and DNA
, used in medicine and in molecular biology research.
Nucleic acid
s are chains of nucleotides, which are composed of three parts: a phosphate
backbone, a pucker-shaped pentose sugar, either ribose
or deoxyribose
, and one of four nucleobase
s.
An analogue may have any of these altered. Typically the analogue nucleobases confer, among other things, different base pairing and base stacking proprieties. Examples include universal bases, which can pair with all four canon bases, and phosphate-sugar backbone analogues such as PNA, which affect the properties of the chain (PNA can even form a triple helix
).
Artificial nucleic acids include peptide nucleic acid (PNA), Morpholino
and locked nucleic acid
(LNA), as well as glycol nucleic acid
(GNA) and threose nucleic acid
(TNA). Each of these is distinguished from naturally occurring DNA or RNA by changes to the backbone of the molecule.
 Nucleic acid analogues are used in molecular biology for several purposes:
Nucleic acid analogues are used in molecular biology for several purposes:
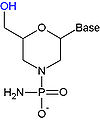 To overcome the fact that ribose
To overcome the fact that ribose
's 2' hydroxy group that reacts with the phosphate linked 3' hydroxy group (RNA is too unstable to be used or synthesized reliably), a ribose analogue is used. The most common RNA analogues are 2'-O-methyl-substituted RNA, locked nucleic acid (LNA
), morpholino
, and peptide nucleic acid (PNA). Although these oligonucleotides have a different backbone sugar or, in the case of PNA, an amino acid residue in place of the ribose phosphate, they still bind to RNA or DNA according to Watson and Crick pairing, but are immune to nuclease activity. They cannot be synthesized enzymatically and can only be obtained synthetically using phosphoramidite strategy
or, for PNA, methods of peptide synthesis
.
are used in sequencing
. These nucleoside triphosphates possess a non-canonical sugar, dideoxyribose, which lacks the 3' hydroxyl group normally present in DNA and therefore cannot bond with the next base. The lack of the 3' hydroxyl group terminates the chain reaction as the DNA polymerases mistake it for a regular deoxyribonucleotide. Another chain-terminating analogue that lacks a 3' hydroxyl and mimics adenosine is called cordycepin
. Cordycepin is an anticancer drug that targets RNA
replication. Another analogue in sequencing is a nucleobase analogue, 7-deaza-GTP and is used to sequence CG rich regions, instead 7-deaza-ATP is called tubercidin, an antibiotic.
several simpler nucleic acids that differ in the backbone, such as TNA
and GNA
and PNA, have been offered as candidates for the first nucleic acids.
(an heterocyclic aromatic six-membered ring with nitrogen atoms in position 1 and 3) and purine
(a pyrimidine (numeration inverted) fused with an imidazole ring, a five-membered ring with 2 nitrogen atoms separated by one carbon (meta), 7,9). Their main proprieties are base pairing, resulting form 2 or 3 hydrogen bonds between ketone (electron withdrawing group
,ei. more negatively charged) and amino (electron releasing group
,ei. more positively charged) functional groups, and base stacking
, caused by the attraction of the delocalized π electron clouds
of the aromatic ring structure.
s (such as rhodamine
or fluorescein
) are linked to the ring linked to the sugar (in para) via a flexible arm, presumably extruding from the major groove of the helix. Due to low processivity of the nucleotides linked to bulky adducts such as florophores by taq polymerases, the sequence is typically copied using a nucleotide with an arm and later coupled with a reactive fluorophore (indirect labelling):
Fluorophores find a variety of uses in medicine and biochemistry.
(Watson-Crick base pairing) via hydrogen bonds (amine with ketone, purine with pyrimidine). Adenine and 2-aminoadenine have one/two amine group(s), whereas thymine has two ketone groups, and cytosine and guanine are mixed amine and ketone (inverted in respect to each other).
The precise reason why there are only four nucleotides is debated, but there are several unused possibilities.
Furthermore, adenine is not the most stable choice for base pairing: in Cyanophage S-2L diaminopurine (DAP) is used instead of adenine (host evasion). Diaminopurine basepairs perfectly with thymine as it is identical to adenine but has an amine group at position 2 forming 3 intramolecular hydrogen bonds, eliminating the major difference between the two types of basepairs (Weak:A-T and Strong:C-G). This improved stability affects protein-binding interactions that rely on those differences.
Other combination include,
However, correct DNA structure can form even when the bases are not paired via hydrogen bonding; that is, the bases pair thanks to hydrophobicity, as studies have shown using DNA isostere
s (analogues with same number of atoms), such as the thymine analogue 2,4-difluorotoluene (F) or the adenine analogue 4-methylbenzimidazole (Z). An alternative hydrophobic pair could be isoquinoline, and the pyrrolo[2,3-b]pyridine
Other noteworthy basepairs:
. Metal-complexing with DNA can occur by the formation of non-canonical base pairs from natural nucleobases with participation by metal ions and also by the exchanging the hydrogen atoms that are part of the Watson-Crick base pairing by metal ions. Introduction of metal ions into a DNA duplex has shown to have potential magnetic, conducting properties, as well as increased stability.
Metal complexing has been shown to occur between natural nucleobase
s. A well-documented example is the formation of T-Hg-T, which involves two deportonated thymine
nucleobases that are brought together by Hg2+ and forms a connected metal-base pair. This motif does not accommodate stacked Hg2+ in a duplex due to an intrastrand hairpin formation process that is favored over duplex formation. Two thymines across from each other in a duplex do not form a Watson-Crick base pair in a duplex; this is an example where a Watson-Crick basepair mismatch is stabilized by the formation of the metal-base pair. Another example of a metal complexing to natural nucleobases is the formation of A-Zn-T and G-Zn-C at high pH; Co+2 and Ni+2 also form these complexes. These are Watson-Crick base pairs where the divalent cation in coordinated to the nucleobases. The exact binding is debated.
A large variety of artificial nucleobases have been developed for use as metal base pairs. These modified nucleobases exhibit tunable electronic properties, sizes, and binding affinities that can be optimized for a specific metal. For, example a nucleoside modified with a pyridine-2,6-dicarboxylate has shown to bind tightly to Cu2+, whereas other divalent ions are only loosely bound. The tridentate character contributes to this selectivity. The fourth coordination site on the copper is saturated by an oppositely arranged pyridine nucleobase. The asymmetric metal base pairing system is orthogonal to the Watson-Crick base pairs. Another example of an artificial nucleobase is that with hydroxypyridone nucleobases, which are able to bind Cu2+ inside the DNA duplex. Five consecutive copper-hydroxypyridone base pairs were incorporated into a double strand, which were flanked by only one natural nucleobase on both ends. EPR data showed that the distance between copper centers was estimated to be 3.7 ± 0.1 Å, while a natural B-type DNA duplex is only slightly larger (3.4 Å). The appeal for stacking metal ions inside a DNA duplex is the hope to obtain nanoscopic self-assembling metal wires, though this has not been realized yet.
Several groups have focused on different aspects:
Analog (chemistry)
In chemistry, a structural analog , also known as chemical analog or simply analog, is a compound having a structure similar to that of another one, but differing from it in respect of a certain component. It can differ in one or more atoms, functional groups, or substructures, which are replaced...
) to naturally occurring RNA
RNA
Ribonucleic acid , or RNA, is one of the three major macromolecules that are essential for all known forms of life....
and DNA
DNA
Deoxyribonucleic acid is a nucleic acid that contains the genetic instructions used in the development and functioning of all known living organisms . The DNA segments that carry this genetic information are called genes, but other DNA sequences have structural purposes, or are involved in...
, used in medicine and in molecular biology research.
Nucleic acid
Nucleic acid
Nucleic acids are biological molecules essential for life, and include DNA and RNA . Together with proteins, nucleic acids make up the most important macromolecules; each is found in abundance in all living things, where they function in encoding, transmitting and expressing genetic information...
s are chains of nucleotides, which are composed of three parts: a phosphate
Phosphate
A phosphate, an inorganic chemical, is a salt of phosphoric acid. In organic chemistry, a phosphate, or organophosphate, is an ester of phosphoric acid. Organic phosphates are important in biochemistry and biogeochemistry or ecology. Inorganic phosphates are mined to obtain phosphorus for use in...
backbone, a pucker-shaped pentose sugar, either ribose
Ribose
Ribose is an organic compound with the formula C5H10O5; specifically, a monosaccharide with linear form H––4–H, which has all the hydroxyl groups on the same side in the Fischer projection....
or deoxyribose
Deoxyribose
Deoxyribose, more, precisely 2-deoxyribose, is a monosaccharide with idealized formula H---3-H. Its name indicates that it is a deoxy sugar, meaning that it is derived from the sugar ribose by loss of an oxygen atom...
, and one of four nucleobase
Nucleobase
Nucleobases are a group of nitrogen-based molecules that are required to form nucleotides, the basic building blocks of DNA and RNA. Nucleobases provide the molecular structure necessary for the hydrogen bonding of complementary DNA and RNA strands, and are key components in the formation of stable...
s.
An analogue may have any of these altered. Typically the analogue nucleobases confer, among other things, different base pairing and base stacking proprieties. Examples include universal bases, which can pair with all four canon bases, and phosphate-sugar backbone analogues such as PNA, which affect the properties of the chain (PNA can even form a triple helix
Triple helix
In geometry, a triple helix is a set of three congruent geometrical helices with the same axis, differing by a translation along the axis. Structures in the form of a triple helix include:* collagen helix...
).
Artificial nucleic acids include peptide nucleic acid (PNA), Morpholino
Morpholino
In molecular biology, a Morpholino is a molecule in a particular structural family that is used to modify gene expression. Morpholino oligomers are an antisense technology used to block access of other molecules to specific sequences within nucleic acid...
and locked nucleic acid
Locked nucleic acid
A locked nucleic acid , often referred to as inaccessible RNA, is a modified RNA nucleotide. The ribose moiety of an LNA nucleotide is modified with an extra bridge connecting the 2' oxygen and 4' carbon. The bridge "locks" the ribose in the 3'-endo conformation, which is often found in the A-form...
(LNA), as well as glycol nucleic acid
GNA (nucleic acid)
Glycol nucleic acid is a polymer similar to DNA or RNA but differing in the composition of its "backbone". GNA is not known to occur naturally; they are synthesized chemically....
(GNA) and threose nucleic acid
TNA (nucleic acid)
Threose nucleic acid is a polymer similar to DNA or RNA but differing in the composition of its "backbone". TNA is not known to occur naturally; they are synthesized chemically....
(TNA). Each of these is distinguished from naturally occurring DNA or RNA by changes to the backbone of the molecule.
Medicine
Several nucleoside analogues are used as antiviral or anticancer agents. The viral polymerase incorporates these compounds with non-canon bases. These compounds are activated in the cells by being converted into nucleotides, they are administered as nucleosides since charged nucleotides cannot easily cross cell membranes.Molecular biology

- As a tool to detect particular sequences
- As a tool with resistance to RNA hydrolysis
- As a tool for another purpose, such as sequencingDNA sequencingDNA sequencing includes several methods and technologies that are used for determining the order of the nucleotide bases—adenine, guanine, cytosine, and thymine—in a molecule of DNA....
- Naturally occurring, such as in tRNA
- Investigation of the mechanisms used by enzyme, such as an Enzyme inhibitorEnzyme inhibitorAn enzyme inhibitor is a molecule that binds to enzymes and decreases their activity. Since blocking an enzyme's activity can kill a pathogen or correct a metabolic imbalance, many drugs are enzyme inhibitors. They are also used as herbicides and pesticides...
- Investigation of possible scenarios of the origin of life
- Investigation of the structural features of nucleic acids
- Investigation of the possible alternatives to the natural system in Synthetic biologySynthetic biologySynthetic biology is a new area of biological research that combines science and engineering. It encompasses a variety of different approaches, methodologies, and disciplines with a variety of definitions...
Hydrolysis resistant RNA-analogues

Ribose
Ribose is an organic compound with the formula C5H10O5; specifically, a monosaccharide with linear form H––4–H, which has all the hydroxyl groups on the same side in the Fischer projection....
's 2' hydroxy group that reacts with the phosphate linked 3' hydroxy group (RNA is too unstable to be used or synthesized reliably), a ribose analogue is used. The most common RNA analogues are 2'-O-methyl-substituted RNA, locked nucleic acid (LNA
Locked nucleic acid
A locked nucleic acid , often referred to as inaccessible RNA, is a modified RNA nucleotide. The ribose moiety of an LNA nucleotide is modified with an extra bridge connecting the 2' oxygen and 4' carbon. The bridge "locks" the ribose in the 3'-endo conformation, which is often found in the A-form...
), morpholino
Morpholino
In molecular biology, a Morpholino is a molecule in a particular structural family that is used to modify gene expression. Morpholino oligomers are an antisense technology used to block access of other molecules to specific sequences within nucleic acid...
, and peptide nucleic acid (PNA). Although these oligonucleotides have a different backbone sugar or, in the case of PNA, an amino acid residue in place of the ribose phosphate, they still bind to RNA or DNA according to Watson and Crick pairing, but are immune to nuclease activity. They cannot be synthesized enzymatically and can only be obtained synthetically using phosphoramidite strategy
Oligonucleotide synthesis
Oligonucleotide synthesis is the chemical synthesis of relatively short fragments of nucleic acids with defined chemical structure . The technique is extremely useful in current laboratory practice because it provides a rapid and inexpensive access to custom-made oligonucleotides of the desired...
or, for PNA, methods of peptide synthesis
Peptide synthesis
In organic chemistry, peptide synthesis is the production of peptides, which are organic compounds in which multiple amino acids are linked via amide bonds which are also known as peptide bonds...
.
Other notable analogues used as tools
DideoxynucleotidesDideoxynucleotides
Dideoxynucleotides, or ddNTPs, are nucleotides lacking a 3'-hydroxyl group on their deoxyribose sugar. Since deoxyribose already lacks a 2'-OH, dideoxyribose lacks hydroxyl groups at both its 2' and 3' carbons...
are used in sequencing
Sequencing
In genetics and biochemistry, sequencing means to determine the primary structure of an unbranched biopolymer...
. These nucleoside triphosphates possess a non-canonical sugar, dideoxyribose, which lacks the 3' hydroxyl group normally present in DNA and therefore cannot bond with the next base. The lack of the 3' hydroxyl group terminates the chain reaction as the DNA polymerases mistake it for a regular deoxyribonucleotide. Another chain-terminating analogue that lacks a 3' hydroxyl and mimics adenosine is called cordycepin
Cordycepin
Cordycepin, or 3'-deoxyadenosine, is a derivative of the nucleoside adenosine, differing from the latter by the absence of oxygen in the 3' position of its ribose part. It was initially extracted from fungi of genus Cordyceps, but is now produced synthetically.Because cordycepin is similar to...
. Cordycepin is an anticancer drug that targets RNA
RNA
Ribonucleic acid , or RNA, is one of the three major macromolecules that are essential for all known forms of life....
replication. Another analogue in sequencing is a nucleobase analogue, 7-deaza-GTP and is used to sequence CG rich regions, instead 7-deaza-ATP is called tubercidin, an antibiotic.
Precursors to the RNA world
RNA may be too complex to be the first nucleic acid, so before the RNA worldRNA world hypothesis
The RNA world hypothesis proposes that life based on ribonucleic acid pre-dates the current world of life based on deoxyribonucleic acid , RNA and proteins. RNA is able both to store genetic information, like DNA, and to catalyze chemical reactions, like an enzyme protein...
several simpler nucleic acids that differ in the backbone, such as TNA
TNA (nucleic acid)
Threose nucleic acid is a polymer similar to DNA or RNA but differing in the composition of its "backbone". TNA is not known to occur naturally; they are synthesized chemically....
and GNA
GNA (nucleic acid)
Glycol nucleic acid is a polymer similar to DNA or RNA but differing in the composition of its "backbone". GNA is not known to occur naturally; they are synthesized chemically....
and PNA, have been offered as candidates for the first nucleic acids.
Nucleobase structure and nomenclature
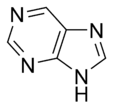
Natural bases are divided into two classes depending on their structure: pyrimidinePyrimidine
Pyrimidine is a heterocyclic aromatic organic compound similar to benzene and pyridine, containing two nitrogen atoms at positions 1 and 3 of the six-member ring...
(an heterocyclic aromatic six-membered ring with nitrogen atoms in position 1 and 3) and purine
Purine
A purine is a heterocyclic aromatic organic compound, consisting of a pyrimidine ring fused to an imidazole ring. Purines, including substituted purines and their tautomers, are the most widely distributed kind of nitrogen-containing heterocycle in nature....
(a pyrimidine (numeration inverted) fused with an imidazole ring, a five-membered ring with 2 nitrogen atoms separated by one carbon (meta), 7,9). Their main proprieties are base pairing, resulting form 2 or 3 hydrogen bonds between ketone (electron withdrawing group
Polar effect
The Polar effect or electronic effect in chemistry is the effect exerted by a substituent on modifying electrostatic forces operating on a nearby reaction center...
,ei. more negatively charged) and amino (electron releasing group
Polar effect
The Polar effect or electronic effect in chemistry is the effect exerted by a substituent on modifying electrostatic forces operating on a nearby reaction center...
,ei. more positively charged) functional groups, and base stacking
Stacking (chemistry)
In chemistry, pi stacking refers to attractive, noncovalent interactions between aromatic rings. These interactions are historically thought to be important in to base stacking of DNA nucleotides, protein folding, template-directed synthesis, materials science, and molecular recognition, although...
, caused by the attraction of the delocalized π electron clouds
Delocalized electron
In chemistry, delocalized electrons are electrons in a molecule, ion or solid metal that are not associated with a single atom or one covalent bond....
of the aromatic ring structure.
 |
 |
| Purine Purine A purine is a heterocyclic aromatic organic compound, consisting of a pyrimidine ring fused to an imidazole ring. Purines, including substituted purines and their tautomers, are the most widely distributed kind of nitrogen-containing heterocycle in nature.... |
Pyrimidine Pyrimidine Pyrimidine is a heterocyclic aromatic organic compound similar to benzene and pyridine, containing two nitrogen atoms at positions 1 and 3 of the six-member ring... |
Fluorophores
Commonly fluorophoreFluorophore
A fluorophore, in analogy to a chromophore, is a component of a molecule which causes a molecule to be fluorescent. It is a functional group in a molecule which will absorb energy of a specific wavelength and re-emit energy at a different wavelength...
s (such as rhodamine
Rhodamine
Rhodamine is a family of related chemical compounds, fluorone dyes. Examples are Rhodamine 6G and Rhodamine B. They are used as a dye and as a dye laser gain medium. They are often used as a tracer dye within water to determine the rate and direction of flow and transport...
or fluorescein
Fluorescein
Fluorescein is a synthetic organic compound available as a dark orange/red powder soluble in water and alcohol. It is widely used as a fluorescent tracer for many applications....
) are linked to the ring linked to the sugar (in para) via a flexible arm, presumably extruding from the major groove of the helix. Due to low processivity of the nucleotides linked to bulky adducts such as florophores by taq polymerases, the sequence is typically copied using a nucleotide with an arm and later coupled with a reactive fluorophore (indirect labelling):
- amine reactive: Aminoallyl nucleotideAminoallyl nucleotideAminoallyl nucleotides are used in post-labeling of nucleic acids to be used in microarrays. These nucleotides are formally known as 5--nucleotides since the aminoallyl group is usually attached to carbon 5 of the pyrimidine ring of uracil and cytosine. They are usually abbreviated as aa-, such as...
contain a primary amine group on a linker that reacts with the amino-reactive dye such as a cyanineCyanineCyanine is a non-systematic name of a synthetic dye family belonging to polymethine group. Cyanines have many uses as fluorescent dyes, particularly in biomedical imaging...
or Alexa Fluor dyes, which contain a reactive leaving group, such as a succinimidyl ester (NHS). (base-pairing amino groups are not affected). - thiol reactive: thiol containing nucleotides reacts with the fluorophore linked to a reactive leaving group, such as a maleimide.
- biotinBiotinBiotin, also known as Vitamin H or Coenzyme R, is a water-soluble B-complex vitamin discovered by Bateman in 1916. It is composed of a ureido ring fused with a tetrahydrothiophene ring. A valeric acid substituent is attached to one of the carbon atoms of the tetrahydrothiophene ring...
linked nucleotides rely on the same indirect labelling principle (+ fluorescent streptavidin) and are used in AffymetrixAffymetrixAffymetrix is a company that manufactures DNA microarrays; it is based in Santa Clara, California, United States. The company was founded by Dr. Stephen Fodor in 1992. It began as a unit in Affymax N.V...
DNAchips.
Fluorophores find a variety of uses in medicine and biochemistry.
Fluorescent base analogues
The most commonly used and commercially available fluorescent base analogue, 2-aminopurine (2-AP), has a high-fluorescence quantum yield free in solution (0.68) that is considerably reduced (appr. 100 times but highly dependent on base sequence) when incorporated into nucleic acids. The emission sensitivity of 2-AP to immediate surroundings is shared by other promising and useful fluorescent base analogues like 3-MI, 6-MI, 6-MAP, pyrrolo-dC (also commercially available), modified and improved derivatives of pyrrolo-dC, furan-modified bases and many other ones (see recent reviews). This sensitivity to the microenvironment have been utilized in studies of e.g. structure and dynamics within both DNA and RNA, dynamics and kinetics of DNA-protein interaction and electron transfer within DNA. A newly developed and very interesting group of fluorescent base analogues that has a fluorescence quantum yield that is nearly insensitive to their immediate surroundings is the tricyclic cytosine family. 1,3-Diaza-2-oxophenothiazine, tC, has a fluorescence quantum yield of approximately 0.2 both in single- and in double-strands irrespective of surrounding bases. Also the oxo-homologue of tC called tCO (both commercially available), 1,3-diaza-2-oxophenoxazine, has a quantum yield of 0.2 in double-stranded systems. However, it is somewhat sensitive to surrounding bases in single-strands (quantum yields of 0.14–0.41). The high and stable quantum yields of these base analogues make them very bright, and, in combination with their good base analogue properties (leaves DNA structure and stability next to unperturbed), they are especially useful in fluorescence anisotropy and FRET measurements, areas where other fluorescent base analogues are less accurate. Also, in the same family of cytosine analogues, a FRET-acceptor base analogue, tCnitro, has been developed. Together with tCO as a FRET-donor this constitutes the first nucleic acid base analogue FRET-pair ever developed. The tC-family has, for example, been used in studies related to polymerase DNA-binding and DNA-polymerization mechanisms.Natural non-canon bases
In a cell, there are several noncanon bases present: CpG islands in DNA (are often methylated), all eukaryotic mRNA (capped with a methyl-7-guanosine), and several bases of rRNAs (are methylated). Often, tRNAs are heavily modified postranscriptionally in order to improve their conformation or base pairing, in particular in/near the anticodon: inosine can base pair with C, U, and even with A, whereas thiouridine (with A) is more specific than uracil (with a purine). Other common tRNA base modifications are pseudouridine (which gives its name to the TΨC loop), dihydrouridine (which does not stack as it is not aromatic), queuosine, wyosine, and so forth. Nevertheless these are all modifications to normal bases and are not placed by a polymerase.Base-pairing
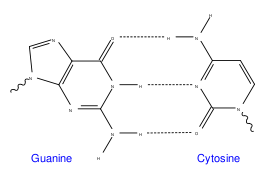
Canonical bases may have either a ketone or an amine group on the carbons surrounding the nitrogen atom furthest away from the glycosidic bond, which allows them to base pairBase pair
In molecular biology and genetics, the linking between two nitrogenous bases on opposite complementary DNA or certain types of RNA strands that are connected via hydrogen bonds is called a base pair...
(Watson-Crick base pairing) via hydrogen bonds (amine with ketone, purine with pyrimidine). Adenine and 2-aminoadenine have one/two amine group(s), whereas thymine has two ketone groups, and cytosine and guanine are mixed amine and ketone (inverted in respect to each other).
| Natural basepairs | |
|---|---|
 |
 |
| A GC basepair: purine ketone/amine forms three intermolecular hydrogen bond Hydrogen bond A hydrogen bond is the attractive interaction of a hydrogen atom with an electronegative atom, such as nitrogen, oxygen or fluorine, that comes from another molecule or chemical group. The hydrogen must be covalently bonded to another electronegative atom to create the bond... s with pyrimidine amine/ketone |
An AT basepair: purine amine/- forms two intermolecular hydrogen bond Hydrogen bond A hydrogen bond is the attractive interaction of a hydrogen atom with an electronegative atom, such as nitrogen, oxygen or fluorine, that comes from another molecule or chemical group. The hydrogen must be covalently bonded to another electronegative atom to create the bond... s with pyrimidine ketone/ketone |
The precise reason why there are only four nucleotides is debated, but there are several unused possibilities.
Furthermore, adenine is not the most stable choice for base pairing: in Cyanophage S-2L diaminopurine (DAP) is used instead of adenine (host evasion). Diaminopurine basepairs perfectly with thymine as it is identical to adenine but has an amine group at position 2 forming 3 intramolecular hydrogen bonds, eliminating the major difference between the two types of basepairs (Weak:A-T and Strong:C-G). This improved stability affects protein-binding interactions that rely on those differences.
Other combination include,
- isoguanine and isocytosine, which have their amine and ketone inverted compared to standard guanine and cytosine, (not used probably as tautomers are problematic for base pairing, but isoC and isoG can be amplified correctly with PCR even in the presence of the 4 canon bases)
- diaminopyrimidine and a xanthine, which bind like 2-aminoadenine and thymine but with inverted structures (not used as xanthine is a deamination product)
| Unused basepair arrangements | ||
|---|---|---|
 |
 |
 |
| A DAP-T base: purine amine/amine forms three intermolecular hydrogen bond Hydrogen bond A hydrogen bond is the attractive interaction of a hydrogen atom with an electronegative atom, such as nitrogen, oxygen or fluorine, that comes from another molecule or chemical group. The hydrogen must be covalently bonded to another electronegative atom to create the bond... s with pyrimidine ketone/ketone |
An X-DAY base: purine ketone/ketone forms three intermolecular hydrogen bond Hydrogen bond A hydrogen bond is the attractive interaction of a hydrogen atom with an electronegative atom, such as nitrogen, oxygen or fluorine, that comes from another molecule or chemical group. The hydrogen must be covalently bonded to another electronegative atom to create the bond... s with pyrimidine amine/amine |
A iG-iC base: purine amine/ketone forms three intermolecular hydrogen bond Hydrogen bond A hydrogen bond is the attractive interaction of a hydrogen atom with an electronegative atom, such as nitrogen, oxygen or fluorine, that comes from another molecule or chemical group. The hydrogen must be covalently bonded to another electronegative atom to create the bond... s with pyrimidine ketone/amine |
However, correct DNA structure can form even when the bases are not paired via hydrogen bonding; that is, the bases pair thanks to hydrophobicity, as studies have shown using DNA isostere
Isostere
Isosteres are molecules or ions with the same number of atoms and the same number of valence electrons.Some examples of isoteres include:*Sodium and hydrogen*Carbon dioxide and nitrous oxide *Silicon and carbon...
s (analogues with same number of atoms), such as the thymine analogue 2,4-difluorotoluene (F) or the adenine analogue 4-methylbenzimidazole (Z). An alternative hydrophobic pair could be isoquinoline, and the pyrrolo[2,3-b]pyridine
Other noteworthy basepairs:
- Several fluorescent bases have also been made, such as the 2-amino-6-(2-thienyl)purine and pyrrole-2-carbaldehyde base pair.
- Metal coordinated bases, such as two 2,6-bis(ethylthiomethyl)pyridine (SPy) with a silver ion or pyridine-2,6-dicarboxamide (Dipam) and a mondentate pyridine (Py) with a copper ion.
- Universal bases may pair indiscriminately with any other base, but, in general, lower the melting temperature of the sequence considerably; examples include 2'-deoxyinosine (hypoxanthine deoxynucleotide) derivatives, nitroazole analogues, and hydrophobic aromatic non-hydrogen-bonding bases (strong stacking effects). These are used as proof of concept and, in general, are not utilised in degenerate primers (which are a mixture of primers).
- The numbers of possible base pairs is doubled when xDNAXDNAxDNA is a modified form of DNA with 8 nucleobases: the four natural bases A, C, G, and T, and four artificial modifications of these made longer by the addition of an extra benzene ring: xA, xC, xG, and xT. A pairs with xT, C pairs with xG, G pairs with xC, and T pairs with xA, so the distance...
is considered. xDNA contains expanded bases, in which a benzene ring has been added, which may pair with canon bases, resulting in four possible base-pairs (8 bases:xA-T,xT-A,xC-G,xG-C, 16 bases if the unused arrangements are used). Another form of benzene added bases is yDNA, in which the base is widened by the benzene.
| Novel basepairs with special proprieties | |||
|---|---|---|---|
 |
 |
 |
|
| A F-Z base: methylbenzimidazole does not form intermolecular hydrogen bond Hydrogen bond A hydrogen bond is the attractive interaction of a hydrogen atom with an electronegative atom, such as nitrogen, oxygen or fluorine, that comes from another molecule or chemical group. The hydrogen must be covalently bonded to another electronegative atom to create the bond... s with toluene F/F |
An S-Pa base: purine thienyl/amine forms three intermolecular hydrogen bond Hydrogen bond A hydrogen bond is the attractive interaction of a hydrogen atom with an electronegative atom, such as nitrogen, oxygen or fluorine, that comes from another molecule or chemical group. The hydrogen must be covalently bonded to another electronegative atom to create the bond... s with pyrrole -/carbaldehyde |
An xA-T base: same bonding as A-T |
Metal Base Pairs
In metal base-pairing, the Watson-Crick hydrogen bonds are replaced by the interaction between a metal ion with nucleosides acting as ligands. The possible geometries of the metal that would allow for duplex formation with two bidentate nucleosides around a central metal atom are: tetrahedral, dodecahedral, and square planarSquare planar
The square planar molecular geometry in chemistry describes the stereochemistry that is adopted by certain chemical compounds...
. Metal-complexing with DNA can occur by the formation of non-canonical base pairs from natural nucleobases with participation by metal ions and also by the exchanging the hydrogen atoms that are part of the Watson-Crick base pairing by metal ions. Introduction of metal ions into a DNA duplex has shown to have potential magnetic, conducting properties, as well as increased stability.
Metal complexing has been shown to occur between natural nucleobase
Nucleobase
Nucleobases are a group of nitrogen-based molecules that are required to form nucleotides, the basic building blocks of DNA and RNA. Nucleobases provide the molecular structure necessary for the hydrogen bonding of complementary DNA and RNA strands, and are key components in the formation of stable...
s. A well-documented example is the formation of T-Hg-T, which involves two deportonated thymine
Thymine
Thymine is one of the four nucleobases in the nucleic acid of DNA that are represented by the letters G–C–A–T. The others are adenine, guanine, and cytosine. Thymine is also known as 5-methyluracil, a pyrimidine nucleobase. As the name suggests, thymine may be derived by methylation of uracil at...
nucleobases that are brought together by Hg2+ and forms a connected metal-base pair. This motif does not accommodate stacked Hg2+ in a duplex due to an intrastrand hairpin formation process that is favored over duplex formation. Two thymines across from each other in a duplex do not form a Watson-Crick base pair in a duplex; this is an example where a Watson-Crick basepair mismatch is stabilized by the formation of the metal-base pair. Another example of a metal complexing to natural nucleobases is the formation of A-Zn-T and G-Zn-C at high pH; Co+2 and Ni+2 also form these complexes. These are Watson-Crick base pairs where the divalent cation in coordinated to the nucleobases. The exact binding is debated.
A large variety of artificial nucleobases have been developed for use as metal base pairs. These modified nucleobases exhibit tunable electronic properties, sizes, and binding affinities that can be optimized for a specific metal. For, example a nucleoside modified with a pyridine-2,6-dicarboxylate has shown to bind tightly to Cu2+, whereas other divalent ions are only loosely bound. The tridentate character contributes to this selectivity. The fourth coordination site on the copper is saturated by an oppositely arranged pyridine nucleobase. The asymmetric metal base pairing system is orthogonal to the Watson-Crick base pairs. Another example of an artificial nucleobase is that with hydroxypyridone nucleobases, which are able to bind Cu2+ inside the DNA duplex. Five consecutive copper-hydroxypyridone base pairs were incorporated into a double strand, which were flanked by only one natural nucleobase on both ends. EPR data showed that the distance between copper centers was estimated to be 3.7 ± 0.1 Å, while a natural B-type DNA duplex is only slightly larger (3.4 Å). The appeal for stacking metal ions inside a DNA duplex is the hope to obtain nanoscopic self-assembling metal wires, though this has not been realized yet.
Orthogonal system
It has been proposed and studied both theoretically and experimentally the possibility of implementing an orthogonal system inside cells independent of the cellular genetic material in order to make a completely safe system, with the possible increase in encoding potentialsSeveral groups have focused on different aspects:
- novel backbones and base pairs as discussed above
- XNA replication/transcription polymerases starting generally from T7 RNA polymerase
- ribosomes (16S16S ribosomal RNA16S ribosomal RNA is a component of the 30S subunit of prokaryotic ribosomes. It is approximately 1.5kb in length...
sequences with altered anti Shine-Dalgarno sequenceShine-Dalgarno sequenceThe Shine-Dalgarno sequence , proposed by Australian scientists John Shine and Lynn Dalgarno , is a ribosomal binding site in the mRNA, generally located 8 basepairs upstream of the start codon AUG. The Shine-Dalgarno sequence exists only in prokaryotes. The six-base consensus sequence is AGGAGG;...
allowing the translation of only orthogonal mRNA with a matching altered Shine-Dalgarno sequence) - novel tRNA encoding non-natural aminoacids. See Expanded genetic codeExpanded genetic codeAn expanded genetic code refers to an artificially modified genetic code in which one or more specific codons have been allocated to encode an amino acid which is not among the twenty/twenty-two found in nature.-Background:...
See also
- Molecular biologyMolecular biologyMolecular biology is the branch of biology that deals with the molecular basis of biological activity. This field overlaps with other areas of biology and chemistry, particularly genetics and biochemistry...
- GeneticsGeneticsGenetics , a discipline of biology, is the science of genes, heredity, and variation in living organisms....
- Synthetic biologySynthetic biologySynthetic biology is a new area of biological research that combines science and engineering. It encompasses a variety of different approaches, methodologies, and disciplines with a variety of definitions...
- Oligonucleotide synthesisOligonucleotide synthesisOligonucleotide synthesis is the chemical synthesis of relatively short fragments of nucleic acids with defined chemical structure . The technique is extremely useful in current laboratory practice because it provides a rapid and inexpensive access to custom-made oligonucleotides of the desired...
- Expanded genetic codeExpanded genetic codeAn expanded genetic code refers to an artificially modified genetic code in which one or more specific codons have been allocated to encode an amino acid which is not among the twenty/twenty-two found in nature.-Background:...
- NucleobaseNucleobaseNucleobases are a group of nitrogen-based molecules that are required to form nucleotides, the basic building blocks of DNA and RNA. Nucleobases provide the molecular structure necessary for the hydrogen bonding of complementary DNA and RNA strands, and are key components in the formation of stable...
, nucleosideNucleosideNucleosides are glycosylamines consisting of a nucleobase bound to a ribose or deoxyribose sugar via a beta-glycosidic linkage...
, nucleotideNucleotideNucleotides are molecules that, when joined together, make up the structural units of RNA and DNA. In addition, nucleotides participate in cellular signaling , and are incorporated into important cofactors of enzymatic reactions...
, and nucleic acidNucleic acidNucleic acids are biological molecules essential for life, and include DNA and RNA . Together with proteins, nucleic acids make up the most important macromolecules; each is found in abundance in all living things, where they function in encoding, transmitting and expressing genetic information... - BiotinBiotinBiotin, also known as Vitamin H or Coenzyme R, is a water-soluble B-complex vitamin discovered by Bateman in 1916. It is composed of a ureido ring fused with a tetrahydrothiophene ring. A valeric acid substituent is attached to one of the carbon atoms of the tetrahydrothiophene ring...
, fluorophoreFluorophoreA fluorophore, in analogy to a chromophore, is a component of a molecule which causes a molecule to be fluorescent. It is a functional group in a molecule which will absorb energy of a specific wavelength and re-emit energy at a different wavelength...
and dark quencherDark quencherA dark quencher is a substance that absorbs excitation energy from a fluorophore and dissipates the energy as heat; while a typical quencher re-emits much of this energy as light . Dark quenchers are used in molecular biology in conjunction with fluorophores... - RibozymeRibozymeA ribozyme is an RNA molecule with a well defined tertiary structure that enables it to catalyze a chemical reaction. Ribozyme means ribonucleic acid enzyme. It may also be called an RNA enzyme or catalytic RNA. Many natural ribozymes catalyze either the hydrolysis of one of their own...

