Type I topoisomerase
Encyclopedia
Type I topoisomerases cut one strand of double-stranded DNA, relax the strand, and reanneal the strands. They are further subdivided into two structurally and mechanistically distinct topoisomerases: type IA and type IB.
Historically, type IA topoisomerases are referred to as prokaryotic topo I, while type IB topoisomerases are referred to as eukaryotic topoisomerase. This distinction, however, no longer applies as type IA and type IB topoisomerases exist in all domains of life.
Functionally, these subclasses perform very specialized functions. Prokaryotic topoisomerase I (topo IA) can only relax negative supercoiled DNA, whereas eukaryotic topoisomerase I (topo IB) can introduce positive supercoils, decatenate single-stranded DNA, and relax DNA.
 Introduction
Introduction
Type IA topoisomerases, historically said to be found in prokaryotes, creates a single break in DNA, and passes a second strand or duplex through the break. This strand passage mechanism shares several features with type IIA topoisomerases. They both form a 5' phosphotyrosine intermediate, and require a divalent metal ion to perform its work. Unlike type II topoisomerases, type IA topoisomerases do not use energy to do its work (with the notable exception of reverse gyrase, see below).
Structure
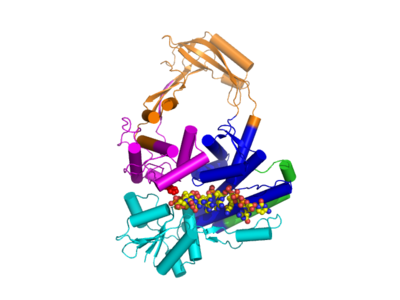
Type IA topoisomerases have several domains, often number Domain 1-4. Domain I contains a Toprim domain (a Rossman fold known to coordinate Magnesium ions), domain IV and domain III each consist of a helix-turn-helix (HTH) domain; the catalytic tyrosine resides on the HTH of domain III. Domain II is a flexible bridge between domains III and IV. The structure of type IA topoisomerase resembles a lock, with Domains I, III and IV lie on the bottom of the structure . The structure of topo III (see below) bound to single-stranded DNA (pdb id = 1I7D) shows how the HTH and Toprim domain are coordinated about the DNA.
Type IA topo variants
There are several variants of Type IA topoisomerases, differing by appendages attached to the main core (sometimes referred to as the "topo-fold"). Members of this subclass include topo I, topo III (which contain additional Zinc-binding motifs), and reverse gyrase. Reverse gyrase is particularly interesting because a ATPase domain, that resembles the helicase-like domain of the Rho transcription factor, is attached (the structure of reverse gyrase was solved by Rodriguez and Stock, EMBO J 2002). The enzyme uses the hydrolysis of ATP to introduce positive supercoils and overwinds DNA, a feature attractive in hyperthermophiles, in which reverse gyrase is known to exist. Rodriguez and Stock have done further work to identify a "latch" that is involved in communicating the hydroylsis of ATP to the introduction of positive supercoils.
The topo III variant is likewise very interesting because it has zinc-binding motifs that is thought to bind single-stranded DNA. Topo III has been identified to be associated with the BLM (for Bloom Syndrome) helicase during recombination.
Mechanism
Type IA topoisomerases operate through a strand-passage mechanism, using a single gate (in contrast with type II topoisomerases). First, the single-stranded DNA binds domain III and I. The catalytic tyrosine cleaves the DNA backbone, creating a transient 5' phosphotyrosine intermediate. The break is then separated, using domain II as a hinge, and a second duplex or strand of DNA is passed through. Domain III and I close and the DNA is re-annealed.
In contrast to type IA topoisomerases, type 1B Topoisomerase solves the problem of overwound and underwound (also referred to as positively or negatively supercoiled) DNA through a hindered rotary mechanism. Crystal structures, biochemistry, and single molecule experiments have contributed to a general mechanism. The enzyme first wraps around DNA
and creates a single, 3' phosphotyrosine intermediate. The 5' end is then free to rotate, twisting it about the other strand, to relax DNA until the topoisomerase religates the broken strands.
Structure
The structure of topo IB bound to DNA has been solved (pdb id = 1A36). Topo IB is composed of an NTD, a capping lobe, a catalytic lobe, and a C-terminal domain. The capping lobe and catalytic lobe wrap around the DNA.
Mechanism
Relaxation is not an active process in the sense that energy (in the form of ATP
) is not spent during the nicking or ligation steps; this is because the reaction between the tyrosine
residue at the active site
of the enzyme with the phosphodiester DNA backbone simply replaces one phosphomonoester bond with another. The topoisomerase also does not use ATP during uncoiling of the DNA; rather, the torque present in the DNA drives the uncoiling and proceeds on average energetically downhill. Recent single molecule experiments have confirmed what bulk-plasmid relaxation experiments have proposed earlier, which is that uncoiling of the DNA is torque-driven and proceeds until religation occurs. No data suggest that Topo IB "controls" the swiveling insofar as that it has a mechanism in place that triggers religation after a specific number of supercoils removed. On the contrary, single-molecule experiments suggest that religation is a random process and has some probability of occurring each time the swiveling 5'-OH end comes in close proximity with the attachment site of the enzyme-linked 3'-end.
Type IB topoisomerases were originally identified in eukaryotes and in viruses. Viral topo I is unique because it binds DNA in a sequence-specific manner.
This intermediate is isoenergetic, meaning that the forward cleavage reaction and the backward religation reaction are both energetically equal. As such, no outside energy source is necessary to conduct this reaction.
Type 1 topoisomerase is inhibited by irinotecan
, topotecan
and camptothecin
.
, named by the association with scleroderma
and the 70 kD extractable immunoreactive fragment that can be obtained from the otherwise larger (100-105 kD) target topoisomerase antigen
(called the SCL-70 Antigen) of the antibodies.
- Type IA topoisomerases change the linking numberLinking numberIn mathematics, the linking number is a numerical invariant that describes the linking of two closed curves in three-dimensional space. Intuitively, the linking number represents the number of times that each curve winds around the other...
of a circular DNA strand by units of strictly 1. - Type IB topoisomerases change the linking number by multiples of 1 (n).
Historically, type IA topoisomerases are referred to as prokaryotic topo I, while type IB topoisomerases are referred to as eukaryotic topoisomerase. This distinction, however, no longer applies as type IA and type IB topoisomerases exist in all domains of life.
Functionally, these subclasses perform very specialized functions. Prokaryotic topoisomerase I (topo IA) can only relax negative supercoiled DNA, whereas eukaryotic topoisomerase I (topo IB) can introduce positive supercoils, decatenate single-stranded DNA, and relax DNA.
Type IA topoisomerses

Type IA topoisomerases, historically said to be found in prokaryotes, creates a single break in DNA, and passes a second strand or duplex through the break. This strand passage mechanism shares several features with type IIA topoisomerases. They both form a 5' phosphotyrosine intermediate, and require a divalent metal ion to perform its work. Unlike type II topoisomerases, type IA topoisomerases do not use energy to do its work (with the notable exception of reverse gyrase, see below).
Structure
Type IA topoisomerases have several domains, often number Domain 1-4. Domain I contains a Toprim domain (a Rossman fold known to coordinate Magnesium ions), domain IV and domain III each consist of a helix-turn-helix (HTH) domain; the catalytic tyrosine resides on the HTH of domain III. Domain II is a flexible bridge between domains III and IV. The structure of type IA topoisomerase resembles a lock, with Domains I, III and IV lie on the bottom of the structure . The structure of topo III (see below) bound to single-stranded DNA (pdb id = 1I7D) shows how the HTH and Toprim domain are coordinated about the DNA.
Type IA topo variants
There are several variants of Type IA topoisomerases, differing by appendages attached to the main core (sometimes referred to as the "topo-fold"). Members of this subclass include topo I, topo III (which contain additional Zinc-binding motifs), and reverse gyrase. Reverse gyrase is particularly interesting because a ATPase domain, that resembles the helicase-like domain of the Rho transcription factor, is attached (the structure of reverse gyrase was solved by Rodriguez and Stock, EMBO J 2002). The enzyme uses the hydrolysis of ATP to introduce positive supercoils and overwinds DNA, a feature attractive in hyperthermophiles, in which reverse gyrase is known to exist. Rodriguez and Stock have done further work to identify a "latch" that is involved in communicating the hydroylsis of ATP to the introduction of positive supercoils.
The topo III variant is likewise very interesting because it has zinc-binding motifs that is thought to bind single-stranded DNA. Topo III has been identified to be associated with the BLM (for Bloom Syndrome) helicase during recombination.
Mechanism
Type IA topoisomerases operate through a strand-passage mechanism, using a single gate (in contrast with type II topoisomerases). First, the single-stranded DNA binds domain III and I. The catalytic tyrosine cleaves the DNA backbone, creating a transient 5' phosphotyrosine intermediate. The break is then separated, using domain II as a hinge, and a second duplex or strand of DNA is passed through. Domain III and I close and the DNA is re-annealed.
Type IB topoisomerases
IntroductionIn contrast to type IA topoisomerases, type 1B Topoisomerase solves the problem of overwound and underwound (also referred to as positively or negatively supercoiled) DNA through a hindered rotary mechanism. Crystal structures, biochemistry, and single molecule experiments have contributed to a general mechanism. The enzyme first wraps around DNA
DNA
Deoxyribonucleic acid is a nucleic acid that contains the genetic instructions used in the development and functioning of all known living organisms . The DNA segments that carry this genetic information are called genes, but other DNA sequences have structural purposes, or are involved in...
and creates a single, 3' phosphotyrosine intermediate. The 5' end is then free to rotate, twisting it about the other strand, to relax DNA until the topoisomerase religates the broken strands.
Structure
The structure of topo IB bound to DNA has been solved (pdb id = 1A36). Topo IB is composed of an NTD, a capping lobe, a catalytic lobe, and a C-terminal domain. The capping lobe and catalytic lobe wrap around the DNA.
Mechanism
Relaxation is not an active process in the sense that energy (in the form of ATP
Adenosine triphosphate
Adenosine-5'-triphosphate is a multifunctional nucleoside triphosphate used in cells as a coenzyme. It is often called the "molecular unit of currency" of intracellular energy transfer. ATP transports chemical energy within cells for metabolism...
) is not spent during the nicking or ligation steps; this is because the reaction between the tyrosine
Tyrosine
Tyrosine or 4-hydroxyphenylalanine, is one of the 22 amino acids that are used by cells to synthesize proteins. Its codons are UAC and UAU. It is a non-essential amino acid with a polar side group...
residue at the active site
Active site
In biology the active site is part of an enzyme where substrates bind and undergo a chemical reaction. The majority of enzymes are proteins but RNA enzymes called ribozymes also exist. The active site of an enzyme is usually found in a cleft or pocket that is lined by amino acid residues that...
of the enzyme with the phosphodiester DNA backbone simply replaces one phosphomonoester bond with another. The topoisomerase also does not use ATP during uncoiling of the DNA; rather, the torque present in the DNA drives the uncoiling and proceeds on average energetically downhill. Recent single molecule experiments have confirmed what bulk-plasmid relaxation experiments have proposed earlier, which is that uncoiling of the DNA is torque-driven and proceeds until religation occurs. No data suggest that Topo IB "controls" the swiveling insofar as that it has a mechanism in place that triggers religation after a specific number of supercoils removed. On the contrary, single-molecule experiments suggest that religation is a random process and has some probability of occurring each time the swiveling 5'-OH end comes in close proximity with the attachment site of the enzyme-linked 3'-end.
Type IB topoisomerases were originally identified in eukaryotes and in viruses. Viral topo I is unique because it binds DNA in a sequence-specific manner.
Type IC topoisomerases
A third type of topoisomerase I was identified, topo V, in the archaeon Methanopyrus kandleri. Topo V is the founding member, and so far the only member, of the type IC topoisomerase, although some authors suggest it may have viral origins . The crystal structure of topo V was solved. Type IC topoisomerases work through a controlled rotary mechanism, much like type IB topoisomerases (pdb ID = 2CSB and 2CSD), but the fold is unique.Intermediates
All topoisomerases form a phosphotyrosine intermediate between the catalytic tyrosine of the enzyme and the scissile phosphoryl of the DNA backbone.- Type IA topoisomerases form a covalent linkage between the catalytic tyrosine and the 5'-phosphoryl.
- Type IB enzymes form a covalent 3'-phosphotyrosine intermediate.
- Type 1C topoisomerases form a covalent 3'-phosphotyrosine intermediate.
This intermediate is isoenergetic, meaning that the forward cleavage reaction and the backward religation reaction are both energetically equal. As such, no outside energy source is necessary to conduct this reaction.
Inhibition
As topoisomerases generate breaks in DNA, they are targets of small-molecule inhibitors that inhibit the enzyme.Type 1 topoisomerase is inhibited by irinotecan
Irinotecan
Irinotecan is a drug used for the treatment of cancer.Irinotecan prevents DNA from unwinding by inhibition of topoisomerase 1. In chemical terms, it is a semisynthetic analogue of the natural alkaloid camptothecin....
, topotecan
Topotecan
Topotecan hydrochloride is a chemotherapy agent that is a topoisomerase I inhibitor. It is the water-soluble derivative of camptothecin...
and camptothecin
Camptothecin
Camptothecin is a cytotoxic quinoline alkaloid which inhibits the DNA enzyme topoisomerase I . It was discovered in 1966 by M. E. Wall and M. C. Wani in systematic screening of natural products for anticancer drugs. It was isolated from the bark and stem of Camptotheca acuminata , a tree native to...
.
Autoantibodies
Autoantibodies targeted against type I topoisomerase are called anti-Scl-70 antibodiesAnti-Scl-70 antibodies
Anti-Scl-70 is an anti-topoisomerase antibody-type of anti-nuclear autoantibodies, seen mainly in diffuse systemic scleroderma , but is also seen in 10–18% of cases of the more limited form of systemic scleroderma called CREST syndrome...
, named by the association with scleroderma
Scleroderma
Systemic sclerosis or systemic scleroderma is a systemic autoimmune disease or systemic connective tissue disease that is a subtype of scleroderma.-Skin symptoms:...
and the 70 kD extractable immunoreactive fragment that can be obtained from the otherwise larger (100-105 kD) target topoisomerase antigen
Antigen
An antigen is a foreign molecule that, when introduced into the body, triggers the production of an antibody by the immune system. The immune system will then kill or neutralize the antigen that is recognized as a foreign and potentially harmful invader. These invaders can be molecules such as...
(called the SCL-70 Antigen) of the antibodies.

