
RNA editing
Encyclopedia
The term RNA editing describes those molecular processes in which the information content in an RNA
molecule is altered through a chemical change in the base makeup. To date, such changes have been observed in tRNA, rRNA, mRNA and microRNA molecules of eukaryotes but not prokaryotes. RNA editing occurs in the cell nucleus
and cytosol
, as well as in mitochondria and plastids, which are thought to have evolved from prokaryotic-like endosymbionts.
Most of the RNA-editing processes, however, appear to be evolutionarily recent acquisitions that arose independently. The diversity of RNA editing mechanisms includes nucleoside modifications such as cytidine
(C) to uridine
(U) and adenosine (A) to inosine
(I) deaminations, as well as non-templated nucleotide additions and insertions. RNA editing in mRNAs effectively alters the amino acid sequence of the encoded protein
so that it differs from that predicted by the genomic DNA sequence
.
has been found in kinetoplasts from the mitochondria of Trypanosoma brucei
Because this may involve a large fraction of the sites in a gene, it is sometimes called "pan-editing" to distinguish it from topical editing of one or a few sites.
Pan-editing starts with the base-pairing of the unedited primary transcript with a guide RNA
(gRNA), which contains complementary sequences to the regions around the insertion/deletion points. The newly formed double-stranded region is then enveloped by an editosome, a large multi-protein complex that catalyzes the editing
. The editosome opens the transcript at the first mismatched nucleotide and starts inserting uridines. The inserted uridines will base-pair with the guide RNA, and insertion will continue as long as A or G is present in the guide RNA and will stop when a C or U is encountered. The inserted nucleotides cause a frameshift and result in a translated protein that differs from its gene. The mechanism of the editosome involves an endonucleolytic cut at the mismatch point between the guide RNA and the unedited transcript. The next step is catalyzed by one of the enzymes in the complex, a terminal U-transferase, which adds Us from UTP at the 3’ end of the mRNA. The opened ends are held in place by other proteins in the complex. Another enzyme, a U-specific exoribonuclease, removes the unpaired Us. After editing has made mRNA complementary to gRNA, an RNA ligase rejoins the ends of the edited mRNA transcript. As a consequence, the editosome can edit only in a 3’ to 5’ direction along the primary RNA transcript. The complex can act on only a single guide RNA at a time. Therefore, a RNA transcript requiring extensive editing will need more than one guide RNA and editosome complex.
The mechanism of the editosome involves an endonucleolytic cut at the mismatch point between the guide RNA and the unedited transcript. The next step is catalyzed by one of the enzymes in the complex, a terminal U-transferase, which adds Us from UTP at the 3’ end of the mRNA. The opened ends are held in place by other proteins in the complex. Another enzyme, a U-specific exoribonuclease, removes the unpaired Us. After editing has made mRNA complementary to gRNA, an RNA ligase rejoins the ends of the edited mRNA transcript. As a consequence, the editosome can edit only in a 3’ to 5’ direction along the primary RNA transcript. The complex can act on only a single guide RNA at a time. Therefore, a RNA transcript requiring extensive editing will need more than one guide RNA and editosome complex.
gene in humans. Apo B100 is expressed in the liver and apo B48 is expressed in the intestines. The B100 form has a CAA sequence that is edited to UAA, a stop codon, in the intestines. It is unedited in the liver.
s) are the RNA-editing enzymes involved in the hydrolytic deamination of Adenosine to Inosine (A-to-I editing). A-to-I editing can be specific (a single adenosine is edited within the stretch of dsRNA) or promiscuous (up to 50% of the adenosines are edited). Specific editing occurs within short duplexes (e.g., those formed in an mRNA where intronic sequence base pairs with a complementary exonic sequence), while promiscuous editing occurs within longer regions of duplex (e.g., pre- or pri-miRNAs, duplexes arising from transgene or viral expression, duplexes arising from paired repetitive elements). There are many effects of A-to-I editing, arising from the fact that I behaves as if it is G both in translation and when forming secondary structures. These effects include alteration of coding capacity, altered miRNA
or siRNA
target populations, heterochromatin formation, nuclear sequestration, cytoplasmic sequestration, endonucleolytic cleavage by Tudor-SN, inhibition of miRNA and siRNA processing ,and altered splicing.
. RNA-editing sites are found mainly in the coding regions of mRNA, introns, and other non-translated regions. In fact, RNA editing can restore the functionality of tRNA molecules. The editing sites are found primarily upstream of mitochondrial or plastid RNAs[39]. The exact mechanism is unknown, but previous studies have speculated the involvement of gRNA and the editosome complex. The reason behind that specific idea arose from the fact that there are too many editing sites that needed to be changed in those organelles for a deaminase.
RNA editing is essential for the normal functioning of the plant’s translation and respiration activity
The term RNA editing describes those molecular processes in which the information content in an RNA
molecule is altered through a chemical change in the base makeup. To date, such changes have been observed in tRNA, rRNA, mRNA and microRNA molecules of eukaryotes but not prokaryotes. RNA editing occurs in the cell nucleus
and cytosol
, as well as in mitochondria and plastids, which are thought to have evolved from prokaryotic-like endosymbionts.
Most of the RNA-editing processes, however, appear to be evolutionarily recent acquisitions that arose independently. The diversity of RNA editing mechanisms includes nucleoside modifications such as cytidine
(C) to uridine
(U) and adenosine (A) to inosine
(I) deaminations, as well as non-templated nucleotide additions and insertions. RNA editing in mRNAs effectively alters the amino acid sequence of the encoded protein
so that it differs from that predicted by the genomic DNA sequence
.
has been found in kinetoplasts from the mitochondria of Trypanosoma brucei
Because this may involve a large fraction of the sites in a gene, it is sometimes called "pan-editing" to distinguish it from topical editing of one or a few sites.
Pan-editing starts with the base-pairing of the unedited primary transcript with a guide RNA
(gRNA), which contains complementary sequences to the regions around the insertion/deletion points. The newly formed double-stranded region is then enveloped by an editosome, a large multi-protein complex that catalyzes the editing
. The editosome opens the transcript at the first mismatched nucleotide and starts inserting uridines. The inserted uridines will base-pair with the guide RNA, and insertion will continue as long as A or G is present in the guide RNA and will stop when a C or U is encountered. The inserted nucleotides cause a frameshift and result in a translated protein that differs from its gene. The mechanism of the editosome involves an endonucleolytic cut at the mismatch point between the guide RNA and the unedited transcript. The next step is catalyzed by one of the enzymes in the complex, a terminal U-transferase, which adds Us from UTP at the 3’ end of the mRNA. The opened ends are held in place by other proteins in the complex. Another enzyme, a U-specific exoribonuclease, removes the unpaired Us. After editing has made mRNA complementary to gRNA, an RNA ligase rejoins the ends of the edited mRNA transcript. As a consequence, the editosome can edit only in a 3’ to 5’ direction along the primary RNA transcript. The complex can act on only a single guide RNA at a time. Therefore, a RNA transcript requiring extensive editing will need more than one guide RNA and editosome complex.
The mechanism of the editosome involves an endonucleolytic cut at the mismatch point between the guide RNA and the unedited transcript. The next step is catalyzed by one of the enzymes in the complex, a terminal U-transferase, which adds Us from UTP at the 3’ end of the mRNA. The opened ends are held in place by other proteins in the complex. Another enzyme, a U-specific exoribonuclease, removes the unpaired Us. After editing has made mRNA complementary to gRNA, an RNA ligase rejoins the ends of the edited mRNA transcript. As a consequence, the editosome can edit only in a 3’ to 5’ direction along the primary RNA transcript. The complex can act on only a single guide RNA at a time. Therefore, a RNA transcript requiring extensive editing will need more than one guide RNA and editosome complex.
gene in humans. Apo B100 is expressed in the liver and apo B48 is expressed in the intestines. The B100 form has a CAA sequence that is edited to UAA, a stop codon, in the intestines. It is unedited in the liver.
s) are the RNA-editing enzymes involved in the hydrolytic deamination of Adenosine to Inosine (A-to-I editing). A-to-I editing can be specific (a single adenosine is edited within the stretch of dsRNA) or promiscuous (up to 50% of the adenosines are edited). Specific editing occurs within short duplexes (e.g., those formed in an mRNA where intronic sequence base pairs with a complementary exonic sequence), while promiscuous editing occurs within longer regions of duplex (e.g., pre- or pri-miRNAs, duplexes arising from transgene or viral expression, duplexes arising from paired repetitive elements). There are many effects of A-to-I editing, arising from the fact that I behaves as if it is G both in translation and when forming secondary structures. These effects include alteration of coding capacity, altered miRNA
or siRNA
target populations, heterochromatin formation, nuclear sequestration, cytoplasmic sequestration, endonucleolytic cleavage by Tudor-SN, inhibition of miRNA and siRNA processing ,and altered splicing.
. RNA-editing sites are found mainly in the coding regions of mRNA, introns, and other non-translated regions. In fact, RNA editing can restore the functionality of tRNA molecules. The editing sites are found primarily upstream of mitochondrial or plastid RNAs[39]. The exact mechanism is unknown, but previous studies have speculated the involvement of gRNA and the editosome complex. The reason behind that specific idea arose from the fact that there are too many editing sites that needed to be changed in those organelles for a deaminase.
RNA editing is essential for the normal functioning of the plant’s translation and respiration activity
The term RNA editing describes those molecular processes in which the information content in an RNA
molecule is altered through a chemical change in the base makeup. To date, such changes have been observed in tRNA, rRNA, mRNA and microRNA molecules of eukaryotes but not prokaryotes. RNA editing occurs in the cell nucleus
and cytosol
, as well as in mitochondria and plastids, which are thought to have evolved from prokaryotic-like endosymbionts.
Most of the RNA-editing processes, however, appear to be evolutionarily recent acquisitions that arose independently. The diversity of RNA editing mechanisms includes nucleoside modifications such as cytidine
(C) to uridine
(U) and adenosine (A) to inosine
(I) deaminations, as well as non-templated nucleotide additions and insertions. RNA editing in mRNAs effectively alters the amino acid sequence of the encoded protein
so that it differs from that predicted by the genomic DNA sequence
.
has been found in kinetoplasts from the mitochondria of Trypanosoma brucei
Because this may involve a large fraction of the sites in a gene, it is sometimes called "pan-editing" to distinguish it from topical editing of one or a few sites.
Pan-editing starts with the base-pairing of the unedited primary transcript with a guide RNA
(gRNA), which contains complementary sequences to the regions around the insertion/deletion points. The newly formed double-stranded region is then enveloped by an editosome, a large multi-protein complex that catalyzes the editing
. The editosome opens the transcript at the first mismatched nucleotide and starts inserting uridines. The inserted uridines will base-pair with the guide RNA, and insertion will continue as long as A or G is present in the guide RNA and will stop when a C or U is encountered. The inserted nucleotides cause a frameshift and result in a translated protein that differs from its gene. The mechanism of the editosome involves an endonucleolytic cut at the mismatch point between the guide RNA and the unedited transcript. The next step is catalyzed by one of the enzymes in the complex, a terminal U-transferase, which adds Us from UTP at the 3’ end of the mRNA. The opened ends are held in place by other proteins in the complex. Another enzyme, a U-specific exoribonuclease, removes the unpaired Us. After editing has made mRNA complementary to gRNA, an RNA ligase rejoins the ends of the edited mRNA transcript. As a consequence, the editosome can edit only in a 3’ to 5’ direction along the primary RNA transcript. The complex can act on only a single guide RNA at a time. Therefore, a RNA transcript requiring extensive editing will need more than one guide RNA and editosome complex.
The mechanism of the editosome involves an endonucleolytic cut at the mismatch point between the guide RNA and the unedited transcript. The next step is catalyzed by one of the enzymes in the complex, a terminal U-transferase, which adds Us from UTP at the 3’ end of the mRNA. The opened ends are held in place by other proteins in the complex. Another enzyme, a U-specific exoribonuclease, removes the unpaired Us. After editing has made mRNA complementary to gRNA, an RNA ligase rejoins the ends of the edited mRNA transcript. As a consequence, the editosome can edit only in a 3’ to 5’ direction along the primary RNA transcript. The complex can act on only a single guide RNA at a time. Therefore, a RNA transcript requiring extensive editing will need more than one guide RNA and editosome complex.
gene in humans. Apo B100 is expressed in the liver and apo B48 is expressed in the intestines. The B100 form has a CAA sequence that is edited to UAA, a stop codon, in the intestines. It is unedited in the liver.
s) are the RNA-editing enzymes involved in the hydrolytic deamination of Adenosine to Inosine (A-to-I editing). A-to-I editing can be specific (a single adenosine is edited within the stretch of dsRNA) or promiscuous (up to 50% of the adenosines are edited). Specific editing occurs within short duplexes (e.g., those formed in an mRNA where intronic sequence base pairs with a complementary exonic sequence), while promiscuous editing occurs within longer regions of duplex (e.g., pre- or pri-miRNAs, duplexes arising from transgene or viral expression, duplexes arising from paired repetitive elements). There are many effects of A-to-I editing, arising from the fact that I behaves as if it is G both in translation and when forming secondary structures. These effects include alteration of coding capacity, altered miRNA
or siRNA
target populations, heterochromatin formation, nuclear sequestration, cytoplasmic sequestration, endonucleolytic cleavage by Tudor-SN, inhibition of miRNA and siRNA processing ,and altered splicing.
. RNA-editing sites are found mainly in the coding regions of mRNA, introns, and other non-translated regions. In fact, RNA editing can restore the functionality of tRNA molecules. The editing sites are found primarily upstream of mitochondrial or plastid RNAs[39]. The exact mechanism is unknown, but previous studies have speculated the involvement of gRNA and the editosome complex. The reason behind that specific idea arose from the fact that there are too many editing sites that needed to be changed in those organelles for a deaminase.
RNA editing is essential for the normal functioning of the plant’s translation and respiration activity It has also been linked to the production of RNA-edited proteins that are incorporated into the polypeptide complexes of the respiration pathway. Therefore, it is highly probable that polypeptides synthesized from unedited RNAs would not function properly and hinder the activity of both mitochondria and plastids.
, mumps
, or parainfluenza) are used for stability and generation of protein variants
. Viral RNAs are transcribed by a virus-encoded RNA-dependent RNA polymerase
, which is prone to pausing and “stuttering” at certain nucleotide combinations. In addition, up to several hundred non-templated As are added by the polymerase at the 3’ end of nascent mRNA. These As help stabilize the mRNA. Furthermore, the pausing and stuttering of the RNA polymerase allows the incorporation of one or two Gs or As upstream of the translational codon. The addition of the non-templated nucleotides shifts the reading frame, which generates a different protein.
, allowing deamination
.
The gRNA-mediated pan-editing in trypanosome mitochondria, involving templated insertion of U residues, is an entirely different biochemical reaction. The enzymes involved have been shown in other studies to be recruited and adapted from different sources. But, the specificity of nucleotide insertion via the interaction between the gRNA and mRNA are similar to the tRNA editing processes in the animal and Acanthamoeba
mithochondria. Eukaryotic ribose methylation of rRNAs by guide RNA molecules is a similar form of modification.
Thus, RNA editing evolved more than once. Several adaptive rationales for editing have been suggested (references in. Editing is often described as a mechanism of correction or repair to compensate for defects in gene sequences. However, in the case of gRNA-mediated editing, this explanation does not seem possible because if a defect happens first, there is no way to generate an error-free gRNA-encoding region, which presumably arises by duplication of the original gene region. This thinking leads to an entirely different evolutionary proposal (called "constructive neutral evolution" in ) in which the order of steps is reversed, with the gratuitous capacity for editing preceding the "defect" 31
RNA
Ribonucleic acid , or RNA, is one of the three major macromolecules that are essential for all known forms of life....
molecule is altered through a chemical change in the base makeup. To date, such changes have been observed in tRNA, rRNA, mRNA and microRNA molecules of eukaryotes but not prokaryotes. RNA editing occurs in the cell nucleus
Cell nucleus
In cell biology, the nucleus is a membrane-enclosed organelle found in eukaryotic cells. It contains most of the cell's genetic material, organized as multiple long linear DNA molecules in complex with a large variety of proteins, such as histones, to form chromosomes. The genes within these...
and cytosol
Cytosol
The cytosol or intracellular fluid is the liquid found inside cells, that is separated into compartments by membranes. For example, the mitochondrial matrix separates the mitochondrion into compartments....
, as well as in mitochondria and plastids, which are thought to have evolved from prokaryotic-like endosymbionts.
Most of the RNA-editing processes, however, appear to be evolutionarily recent acquisitions that arose independently. The diversity of RNA editing mechanisms includes nucleoside modifications such as cytidine
Cytidine
Cytidine is a nucleoside molecule that is formed when cytosine is attached to a ribose ring via a β-N1-glycosidic bond...
(C) to uridine
Uridine
Uridine is a molecule that is formed when uracil is attached to a ribose ring via a β-N1-glycosidic bond.If uracil is attached to a deoxyribose ring, it is known as a deoxyuridine....
(U) and adenosine (A) to inosine
Inosine
Inosine is a nucleoside that is formed when hypoxanthine is attached to a ribose ring via a β-N9-glycosidic bond....
(I) deaminations, as well as non-templated nucleotide additions and insertions. RNA editing in mRNAs effectively alters the amino acid sequence of the encoded protein
Protein
Proteins are biochemical compounds consisting of one or more polypeptides typically folded into a globular or fibrous form, facilitating a biological function. A polypeptide is a single linear polymer chain of amino acids bonded together by peptide bonds between the carboxyl and amino groups of...
so that it differs from that predicted by the genomic DNA sequence
DNA sequence
The sequence or primary structure of a nucleic acid is the composition of atoms that make up the nucleic acid and the chemical bonds that bond those atoms. Because nucleic acids, such as DNA and RNA, are unbranched polymers, this specification is equivalent to specifying the sequence of...
.
Editing by insertion or deletion
RNA editing through the addition and deletion of uracilUracil
Uracil is one of the four nucleobases in the nucleic acid of RNA that are represented by the letters A, G, C and U. The others are adenine, cytosine, and guanine. In RNA, uracil binds to adenine via two hydrogen bonds. In DNA, the uracil nucleobase is replaced by thymine.Uracil is a common and...
has been found in kinetoplasts from the mitochondria of Trypanosoma brucei
Trypanosoma brucei
Trypanosoma brucei is a parasitic protist species that causes African trypanosomiasis in humans and nagana in animals in Africa. There are 3 sub-species of T. brucei: T. b. brucei, T. b. gambiense and T. b. rhodesiense.These obligate parasites have two hosts - an insect vector and mammalian host...
Because this may involve a large fraction of the sites in a gene, it is sometimes called "pan-editing" to distinguish it from topical editing of one or a few sites.
Pan-editing starts with the base-pairing of the unedited primary transcript with a guide RNA
Guide RNA
Guide RNAs are the RNAs that guide the insertion or deletion of uridine residues into mitochondrial mRNAs in kinetoplastid protists in a process known as RNA editing.-Overview of gRNA-directed editing:...
(gRNA), which contains complementary sequences to the regions around the insertion/deletion points. The newly formed double-stranded region is then enveloped by an editosome, a large multi-protein complex that catalyzes the editing
. The editosome opens the transcript at the first mismatched nucleotide and starts inserting uridines. The inserted uridines will base-pair with the guide RNA, and insertion will continue as long as A or G is present in the guide RNA and will stop when a C or U is encountered. The inserted nucleotides cause a frameshift and result in a translated protein that differs from its gene.
C-U editing
The editing involves cytidine deaminase that deaminates a cytidine base into a uridine base. An example of C-to-U editing is with the apolipoprotein BApolipoprotein B
Apolipoprotein B is the primary apolipoprotein of low-density lipoproteins , which is responsible for carrying cholesterol to tissues. While it is unclear exactly what functional role APOB plays in LDL, it is the primary apolipoprotein component and is absolutely required for its formation...
gene in humans. Apo B100 is expressed in the liver and apo B48 is expressed in the intestines. The B100 form has a CAA sequence that is edited to UAA, a stop codon, in the intestines. It is unedited in the liver.
A-I editing
A-to-I editing occurs in regions of double-stranded RNA (dsRNA). Adenosine deaminases acting on RNA (ADARADAR
Double-stranded RNA-specific adenosine deaminase is an enzyme that in humans is encoded by the ADAR gene.-Further reading:...
s) are the RNA-editing enzymes involved in the hydrolytic deamination of Adenosine to Inosine (A-to-I editing). A-to-I editing can be specific (a single adenosine is edited within the stretch of dsRNA) or promiscuous (up to 50% of the adenosines are edited). Specific editing occurs within short duplexes (e.g., those formed in an mRNA where intronic sequence base pairs with a complementary exonic sequence), while promiscuous editing occurs within longer regions of duplex (e.g., pre- or pri-miRNAs, duplexes arising from transgene or viral expression, duplexes arising from paired repetitive elements). There are many effects of A-to-I editing, arising from the fact that I behaves as if it is G both in translation and when forming secondary structures. These effects include alteration of coding capacity, altered miRNA
Mirna
Mirna may refer to:geographical entities* Mirna , a river in Istria, Croatia* Mirna , a river in Slovenia, tributary of the river Sava* Mirna , a settlement in the municipality of Mirna in Southeastern Sloveniapeople...
or siRNA
Sírna
Sírna Sáeglach , son of Dian mac Demal, son of Demal mac Rothechtaid, son of Rothechtaid mac Main, was, according to medieval Irish legend and historical tradition, a High King of Ireland...
target populations, heterochromatin formation, nuclear sequestration, cytoplasmic sequestration, endonucleolytic cleavage by Tudor-SN, inhibition of miRNA and siRNA processing ,and altered splicing.
RNA editing in plant mitochondria and plastids
It has been shown in previous studies that the only types of RNA editing seen in the plants’ mitochondria and plastids are conversion of C to U and U to C (very rare). RNA-editing sites are found mainly in the coding regions of mRNA, introns, and other non-translated regions. In fact, RNA editing can restore the functionality of tRNA molecules. The editing sites are found primarily upstream of mitochondrial or plastid RNAs[39]. The exact mechanism is unknown, but previous studies have speculated the involvement of gRNA and the editosome complex. The reason behind that specific idea arose from the fact that there are too many editing sites that needed to be changed in those organelles for a deaminase.
RNA editing is essential for the normal functioning of the plant’s translation and respiration activity
The term RNA editing describes those molecular processes in which the information content in an RNA
RNA
Ribonucleic acid , or RNA, is one of the three major macromolecules that are essential for all known forms of life....
molecule is altered through a chemical change in the base makeup. To date, such changes have been observed in tRNA, rRNA, mRNA and microRNA molecules of eukaryotes but not prokaryotes. RNA editing occurs in the cell nucleus
Cell nucleus
In cell biology, the nucleus is a membrane-enclosed organelle found in eukaryotic cells. It contains most of the cell's genetic material, organized as multiple long linear DNA molecules in complex with a large variety of proteins, such as histones, to form chromosomes. The genes within these...
and cytosol
Cytosol
The cytosol or intracellular fluid is the liquid found inside cells, that is separated into compartments by membranes. For example, the mitochondrial matrix separates the mitochondrion into compartments....
, as well as in mitochondria and plastids, which are thought to have evolved from prokaryotic-like endosymbionts.
Most of the RNA-editing processes, however, appear to be evolutionarily recent acquisitions that arose independently. The diversity of RNA editing mechanisms includes nucleoside modifications such as cytidine
Cytidine
Cytidine is a nucleoside molecule that is formed when cytosine is attached to a ribose ring via a β-N1-glycosidic bond...
(C) to uridine
Uridine
Uridine is a molecule that is formed when uracil is attached to a ribose ring via a β-N1-glycosidic bond.If uracil is attached to a deoxyribose ring, it is known as a deoxyuridine....
(U) and adenosine (A) to inosine
Inosine
Inosine is a nucleoside that is formed when hypoxanthine is attached to a ribose ring via a β-N9-glycosidic bond....
(I) deaminations, as well as non-templated nucleotide additions and insertions. RNA editing in mRNAs effectively alters the amino acid sequence of the encoded protein
Protein
Proteins are biochemical compounds consisting of one or more polypeptides typically folded into a globular or fibrous form, facilitating a biological function. A polypeptide is a single linear polymer chain of amino acids bonded together by peptide bonds between the carboxyl and amino groups of...
so that it differs from that predicted by the genomic DNA sequence
DNA sequence
The sequence or primary structure of a nucleic acid is the composition of atoms that make up the nucleic acid and the chemical bonds that bond those atoms. Because nucleic acids, such as DNA and RNA, are unbranched polymers, this specification is equivalent to specifying the sequence of...
.
Editing by insertion or deletion
RNA editing through the addition and deletion of uracilUracil
Uracil is one of the four nucleobases in the nucleic acid of RNA that are represented by the letters A, G, C and U. The others are adenine, cytosine, and guanine. In RNA, uracil binds to adenine via two hydrogen bonds. In DNA, the uracil nucleobase is replaced by thymine.Uracil is a common and...
has been found in kinetoplasts from the mitochondria of Trypanosoma brucei
Trypanosoma brucei
Trypanosoma brucei is a parasitic protist species that causes African trypanosomiasis in humans and nagana in animals in Africa. There are 3 sub-species of T. brucei: T. b. brucei, T. b. gambiense and T. b. rhodesiense.These obligate parasites have two hosts - an insect vector and mammalian host...
Because this may involve a large fraction of the sites in a gene, it is sometimes called "pan-editing" to distinguish it from topical editing of one or a few sites.
Pan-editing starts with the base-pairing of the unedited primary transcript with a guide RNA
Guide RNA
Guide RNAs are the RNAs that guide the insertion or deletion of uridine residues into mitochondrial mRNAs in kinetoplastid protists in a process known as RNA editing.-Overview of gRNA-directed editing:...
(gRNA), which contains complementary sequences to the regions around the insertion/deletion points. The newly formed double-stranded region is then enveloped by an editosome, a large multi-protein complex that catalyzes the editing
. The editosome opens the transcript at the first mismatched nucleotide and starts inserting uridines. The inserted uridines will base-pair with the guide RNA, and insertion will continue as long as A or G is present in the guide RNA and will stop when a C or U is encountered. The inserted nucleotides cause a frameshift and result in a translated protein that differs from its gene.
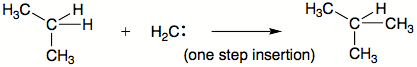
C-U editing
The editing involves cytidine deaminase that deaminates a cytidine base into a uridine base. An example of C-to-U editing is with the apolipoprotein BApolipoprotein B
Apolipoprotein B is the primary apolipoprotein of low-density lipoproteins , which is responsible for carrying cholesterol to tissues. While it is unclear exactly what functional role APOB plays in LDL, it is the primary apolipoprotein component and is absolutely required for its formation...
gene in humans. Apo B100 is expressed in the liver and apo B48 is expressed in the intestines. The B100 form has a CAA sequence that is edited to UAA, a stop codon, in the intestines. It is unedited in the liver.
A-I editing
A-to-I editing occurs in regions of double-stranded RNA (dsRNA). Adenosine deaminases acting on RNA (ADARADAR
Double-stranded RNA-specific adenosine deaminase is an enzyme that in humans is encoded by the ADAR gene.-Further reading:...
s) are the RNA-editing enzymes involved in the hydrolytic deamination of Adenosine to Inosine (A-to-I editing). A-to-I editing can be specific (a single adenosine is edited within the stretch of dsRNA) or promiscuous (up to 50% of the adenosines are edited). Specific editing occurs within short duplexes (e.g., those formed in an mRNA where intronic sequence base pairs with a complementary exonic sequence), while promiscuous editing occurs within longer regions of duplex (e.g., pre- or pri-miRNAs, duplexes arising from transgene or viral expression, duplexes arising from paired repetitive elements). There are many effects of A-to-I editing, arising from the fact that I behaves as if it is G both in translation and when forming secondary structures. These effects include alteration of coding capacity, altered miRNA
Mirna
Mirna may refer to:geographical entities* Mirna , a river in Istria, Croatia* Mirna , a river in Slovenia, tributary of the river Sava* Mirna , a settlement in the municipality of Mirna in Southeastern Sloveniapeople...
or siRNA
Sírna
Sírna Sáeglach , son of Dian mac Demal, son of Demal mac Rothechtaid, son of Rothechtaid mac Main, was, according to medieval Irish legend and historical tradition, a High King of Ireland...
target populations, heterochromatin formation, nuclear sequestration, cytoplasmic sequestration, endonucleolytic cleavage by Tudor-SN, inhibition of miRNA and siRNA processing ,and altered splicing.
RNA editing in plant mitochondria and plastids
It has been shown in previous studies that the only types of RNA editing seen in the plants’ mitochondria and plastids are conversion of C to U and U to C (very rare). RNA-editing sites are found mainly in the coding regions of mRNA, introns, and other non-translated regions. In fact, RNA editing can restore the functionality of tRNA molecules. The editing sites are found primarily upstream of mitochondrial or plastid RNAs[39]. The exact mechanism is unknown, but previous studies have speculated the involvement of gRNA and the editosome complex. The reason behind that specific idea arose from the fact that there are too many editing sites that needed to be changed in those organelles for a deaminase.
RNA editing is essential for the normal functioning of the plant’s translation and respiration activity
The term RNA editing describes those molecular processes in which the information content in an RNA
RNA
Ribonucleic acid , or RNA, is one of the three major macromolecules that are essential for all known forms of life....
molecule is altered through a chemical change in the base makeup. To date, such changes have been observed in tRNA, rRNA, mRNA and microRNA molecules of eukaryotes but not prokaryotes. RNA editing occurs in the cell nucleus
Cell nucleus
In cell biology, the nucleus is a membrane-enclosed organelle found in eukaryotic cells. It contains most of the cell's genetic material, organized as multiple long linear DNA molecules in complex with a large variety of proteins, such as histones, to form chromosomes. The genes within these...
and cytosol
Cytosol
The cytosol or intracellular fluid is the liquid found inside cells, that is separated into compartments by membranes. For example, the mitochondrial matrix separates the mitochondrion into compartments....
, as well as in mitochondria and plastids, which are thought to have evolved from prokaryotic-like endosymbionts.
Most of the RNA-editing processes, however, appear to be evolutionarily recent acquisitions that arose independently. The diversity of RNA editing mechanisms includes nucleoside modifications such as cytidine
Cytidine
Cytidine is a nucleoside molecule that is formed when cytosine is attached to a ribose ring via a β-N1-glycosidic bond...
(C) to uridine
Uridine
Uridine is a molecule that is formed when uracil is attached to a ribose ring via a β-N1-glycosidic bond.If uracil is attached to a deoxyribose ring, it is known as a deoxyuridine....
(U) and adenosine (A) to inosine
Inosine
Inosine is a nucleoside that is formed when hypoxanthine is attached to a ribose ring via a β-N9-glycosidic bond....
(I) deaminations, as well as non-templated nucleotide additions and insertions. RNA editing in mRNAs effectively alters the amino acid sequence of the encoded protein
Protein
Proteins are biochemical compounds consisting of one or more polypeptides typically folded into a globular or fibrous form, facilitating a biological function. A polypeptide is a single linear polymer chain of amino acids bonded together by peptide bonds between the carboxyl and amino groups of...
so that it differs from that predicted by the genomic DNA sequence
DNA sequence
The sequence or primary structure of a nucleic acid is the composition of atoms that make up the nucleic acid and the chemical bonds that bond those atoms. Because nucleic acids, such as DNA and RNA, are unbranched polymers, this specification is equivalent to specifying the sequence of...
.
Editing by insertion or deletion
RNA editing through the addition and deletion of uracilUracil
Uracil is one of the four nucleobases in the nucleic acid of RNA that are represented by the letters A, G, C and U. The others are adenine, cytosine, and guanine. In RNA, uracil binds to adenine via two hydrogen bonds. In DNA, the uracil nucleobase is replaced by thymine.Uracil is a common and...
has been found in kinetoplasts from the mitochondria of Trypanosoma brucei
Trypanosoma brucei
Trypanosoma brucei is a parasitic protist species that causes African trypanosomiasis in humans and nagana in animals in Africa. There are 3 sub-species of T. brucei: T. b. brucei, T. b. gambiense and T. b. rhodesiense.These obligate parasites have two hosts - an insect vector and mammalian host...
Because this may involve a large fraction of the sites in a gene, it is sometimes called "pan-editing" to distinguish it from topical editing of one or a few sites.
Pan-editing starts with the base-pairing of the unedited primary transcript with a guide RNA
Guide RNA
Guide RNAs are the RNAs that guide the insertion or deletion of uridine residues into mitochondrial mRNAs in kinetoplastid protists in a process known as RNA editing.-Overview of gRNA-directed editing:...
(gRNA), which contains complementary sequences to the regions around the insertion/deletion points. The newly formed double-stranded region is then enveloped by an editosome, a large multi-protein complex that catalyzes the editing
. The editosome opens the transcript at the first mismatched nucleotide and starts inserting uridines. The inserted uridines will base-pair with the guide RNA, and insertion will continue as long as A or G is present in the guide RNA and will stop when a C or U is encountered. The inserted nucleotides cause a frameshift and result in a translated protein that differs from its gene.
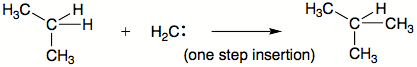
C-U editing
The editing involves cytidine deaminase that deaminates a cytidine base into a uridine base. An example of C-to-U editing is with the apolipoprotein BApolipoprotein B
Apolipoprotein B is the primary apolipoprotein of low-density lipoproteins , which is responsible for carrying cholesterol to tissues. While it is unclear exactly what functional role APOB plays in LDL, it is the primary apolipoprotein component and is absolutely required for its formation...
gene in humans. Apo B100 is expressed in the liver and apo B48 is expressed in the intestines. The B100 form has a CAA sequence that is edited to UAA, a stop codon, in the intestines. It is unedited in the liver.
A-I editing
A-to-I editing occurs in regions of double-stranded RNA (dsRNA). Adenosine deaminases acting on RNA (ADARADAR
Double-stranded RNA-specific adenosine deaminase is an enzyme that in humans is encoded by the ADAR gene.-Further reading:...
s) are the RNA-editing enzymes involved in the hydrolytic deamination of Adenosine to Inosine (A-to-I editing). A-to-I editing can be specific (a single adenosine is edited within the stretch of dsRNA) or promiscuous (up to 50% of the adenosines are edited). Specific editing occurs within short duplexes (e.g., those formed in an mRNA where intronic sequence base pairs with a complementary exonic sequence), while promiscuous editing occurs within longer regions of duplex (e.g., pre- or pri-miRNAs, duplexes arising from transgene or viral expression, duplexes arising from paired repetitive elements). There are many effects of A-to-I editing, arising from the fact that I behaves as if it is G both in translation and when forming secondary structures. These effects include alteration of coding capacity, altered miRNA
Mirna
Mirna may refer to:geographical entities* Mirna , a river in Istria, Croatia* Mirna , a river in Slovenia, tributary of the river Sava* Mirna , a settlement in the municipality of Mirna in Southeastern Sloveniapeople...
or siRNA
Sírna
Sírna Sáeglach , son of Dian mac Demal, son of Demal mac Rothechtaid, son of Rothechtaid mac Main, was, according to medieval Irish legend and historical tradition, a High King of Ireland...
target populations, heterochromatin formation, nuclear sequestration, cytoplasmic sequestration, endonucleolytic cleavage by Tudor-SN, inhibition of miRNA and siRNA processing ,and altered splicing.
RNA editing in plant mitochondria and plastids
It has been shown in previous studies that the only types of RNA editing seen in the plants’ mitochondria and plastids are conversion of C to U and U to C (very rare). RNA-editing sites are found mainly in the coding regions of mRNA, introns, and other non-translated regions. In fact, RNA editing can restore the functionality of tRNA molecules. The editing sites are found primarily upstream of mitochondrial or plastid RNAs[39]. The exact mechanism is unknown, but previous studies have speculated the involvement of gRNA and the editosome complex. The reason behind that specific idea arose from the fact that there are too many editing sites that needed to be changed in those organelles for a deaminase.
RNA editing is essential for the normal functioning of the plant’s translation and respiration activity It has also been linked to the production of RNA-edited proteins that are incorporated into the polypeptide complexes of the respiration pathway. Therefore, it is highly probable that polypeptides synthesized from unedited RNAs would not function properly and hinder the activity of both mitochondria and plastids.
RNA editing in viruses
RNA editing in viruses (i.e., measlesMeasles
Measles, also known as rubeola or morbilli, is an infection of the respiratory system caused by a virus, specifically a paramyxovirus of the genus Morbillivirus. Morbilliviruses, like other paramyxoviruses, are enveloped, single-stranded, negative-sense RNA viruses...
, mumps
Mumps
Mumps is a viral disease of the human species, caused by the mumps virus. Before the development of vaccination and the introduction of a vaccine, it was a common childhood disease worldwide...
, or parainfluenza) are used for stability and generation of protein variants
. Viral RNAs are transcribed by a virus-encoded RNA-dependent RNA polymerase
RNA polymerase
RNA polymerase is an enzyme that produces RNA. In cells, RNAP is needed for constructing RNA chains from DNA genes as templates, a process called transcription. RNA polymerase enzymes are essential to life and are found in all organisms and many viruses...
, which is prone to pausing and “stuttering” at certain nucleotide combinations. In addition, up to several hundred non-templated As are added by the polymerase at the 3’ end of nascent mRNA. These As help stabilize the mRNA. Furthermore, the pausing and stuttering of the RNA polymerase allows the incorporation of one or two Gs or As upstream of the translational codon. The addition of the non-templated nucleotides shifts the reading frame, which generates a different protein.
Origin and evolution of RNA editing
The RNA-editing system seen in the animal may have evolved from mononucleotide deaminases, which have led to larger gene families that include the apobec-1 and adar genes. These genes share close identity with the bacterial deaminases involved in nucleotide metabolism. The adenosine deaminase of E. coli cannot deaminate a nucleoside in the RNA; the enzyme’s reaction pocket is too small to for the RNA strand to bind to. However, this active site is widened by amino acid changes in the corresponding human analog genes, APOBEC-1 and ADARADAR
Double-stranded RNA-specific adenosine deaminase is an enzyme that in humans is encoded by the ADAR gene.-Further reading:...
, allowing deamination
.
The gRNA-mediated pan-editing in trypanosome mitochondria, involving templated insertion of U residues, is an entirely different biochemical reaction. The enzymes involved have been shown in other studies to be recruited and adapted from different sources. But, the specificity of nucleotide insertion via the interaction between the gRNA and mRNA are similar to the tRNA editing processes in the animal and Acanthamoeba
Acanthamoeba
Acanthamoeba is a genus of amoebae, one of the most common protozoa in soil, and also frequently found in fresh water and other habitats. The cells are small, usually 15 to 35 μm in length and oval to triangular in shape when moving. The pseudopods form a clear hemispherical lobe at the anterior,...
mithochondria. Eukaryotic ribose methylation of rRNAs by guide RNA molecules is a similar form of modification.
Thus, RNA editing evolved more than once. Several adaptive rationales for editing have been suggested (references in. Editing is often described as a mechanism of correction or repair to compensate for defects in gene sequences. However, in the case of gRNA-mediated editing, this explanation does not seem possible because if a defect happens first, there is no way to generate an error-free gRNA-encoding region, which presumably arises by duplication of the original gene region. This thinking leads to an entirely different evolutionary proposal (called "constructive neutral evolution" in ) in which the order of steps is reversed, with the gratuitous capacity for editing preceding the "defect" 31

