Archaeal Richmond Mine Acidophilic Nanoorganisms
Encyclopedia
Archaeal Richmond Mine acidophilic nanoorganisms (ARMAN) were first discovered in an extremely acidic mine located in northern California (Iron Mountain Mine
) by Brett Baker in Jill Banfield's laboratory at the University of California Berkeley. These novel groups of archaea
named ARMAN-1, ARMAN-2 (Candidatus Micrarchaeum acidiphilum ARMAN-2 ), and ARMAN-3 were missed by previous PCR-based surveys of the mine community because the ARMANs have several mismatches with commonly used PCR primers for 16S rRNA genes. Baker et al. detected them in a later study using shotgun sequencing
of the community. The three groups represent three unique lineages deeply branched within the Euryarchaeota
, a subgroup of the Archaea
. Their 16S rRNA genes differ by as much as 17% between the three groups. Prior to their discovery all of the Archaea
shown to be associated with Iron Mountain belonged to the order Thermoplasmatales
(e.g., Ferroplasma
acidarmanus).
(AMD), at Iron Mountain in northern California, that have pH < 1.5. They are usually found in low abundance (5–25%) in the community. More recently, closely related organisms have been detected in an acidic boreal mire or bog
in Finland and another acid mine drainage site in extreme environments of Rio Tinto, southwestern Spain.
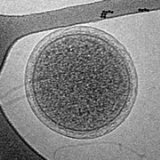
an extensive 3D characterization of uncultivated ARMAN cells within mine biofilms has been done (Comolli et al. 2009). This has revealed that they are right at the cell size predicted to be the lower limit for life, 0.009 µm3 and 0.04 µm3 (NRC Steering group). They also found that despite their unusually small cell size it is common to find more than one type of virus
attached to the cells while in the biofilms. Furthermore, the cells contain on average ~92 ribosomes per cell, whereas the average E. coli cell grown in culture contains ~10,000 ribosomes. This suggests that for ARMAN cells a much more limited number of metabolites are present in a given cell. It raises questions about what the minimal requirements are for a living cell.
3D reconstructions of ARMAN cells in the environment has revealed that a small number of them attach to other Archaea of the order Thermoplasmatales
(Baker et al. 2010 ). The ARMAN cells appear to penetrate the cell wall to the cytoplasm of the Thermoplasmatales cells. The nature of this interaction hasn't been determined. It could be some sort of parasitic or symbiotic interaction. It's possible that ARMAN is getting some sort of metabolite that it is not able to produce on its own.
during a 2006 Community Sequencing Program (CSP). These three genomes were successfully binned from the community genomic data using ESOM or Emergent Self-Organizing Map
clustering of tetra-nucleotide DNA signatures (Dick et al. 2009).
The first draft of Candidatus Micrarchaeum acidiphilum ARMAN-2 is ~1 Mb (3 scaffolds, Baker et al.. PNAS 2010. The ARMAN-2 has recently been closed using 454 and Solexa sequencing of other biofilms to close the gaps and is being prepared for submission to NCBI. The genomes of ARMAN-4 and ARMAN-5 (roughly 1 Mb as well) have unusually average genes sizes, similar to those seen in endosymbiotic and parasitic bacteria. This may be signature of their inter-species interactions with other Archaea in nature (Baker et al. 2010 ). Furthermore, the branching of these groups near the Euryarchaea/Crenarchaea divide is reflected in them sharing many genetic aspects of both Crenarchaea and Euryarchaea. Specifically they have many genes that had previously only been identified in Crenarchaea. It is difficult to elucidate many of the commonly known metabolic pathways in ARMAN due to the unusually high number of unique genes that have been identified in their genomes.
A novel type of tRNA splicing endonuclease, involved in the processing of tRNA, has been discovered in ARMAN groups 1 and 2 (Fujishima et al. 2011). The enzyme consists of two duplicated catalytic units and one structural unit encoded on a single gene, representing a novel three-unit architecture.
Iron Mountain Mine
Iron Mountain Mine, also known as the Richmond Mine at Iron Mountain, is a mine near Redding in Northern California. Geologically classified as a "massive sulfide ore deposit", the site was mined for iron, silver, gold, copper, zinc, and pyrite intermittently from the 1860s until 1963...
) by Brett Baker in Jill Banfield's laboratory at the University of California Berkeley. These novel groups of archaea
Archaea
The Archaea are a group of single-celled microorganisms. A single individual or species from this domain is called an archaeon...
named ARMAN-1, ARMAN-2 (Candidatus Micrarchaeum acidiphilum ARMAN-2 ), and ARMAN-3 were missed by previous PCR-based surveys of the mine community because the ARMANs have several mismatches with commonly used PCR primers for 16S rRNA genes. Baker et al. detected them in a later study using shotgun sequencing
Shotgun sequencing
In genetics, shotgun sequencing, also known as shotgun cloning, is a method used for sequencing long DNA strands. It is named by analogy with the rapidly-expanding, quasi-random firing pattern of a shotgun....
of the community. The three groups represent three unique lineages deeply branched within the Euryarchaeota
Euryarchaeota
In the taxonomy of microorganisms, the Euryarchaeota are a phylum of the Archaea.The Euryarchaeota include the methanogens, which produce methane and are often found in intestines, the halobacteria, which survive extreme concentrations of salt, and some extremely thermophilic aerobes and anaerobes...
, a subgroup of the Archaea
Archaea
The Archaea are a group of single-celled microorganisms. A single individual or species from this domain is called an archaeon...
. Their 16S rRNA genes differ by as much as 17% between the three groups. Prior to their discovery all of the Archaea
Archaea
The Archaea are a group of single-celled microorganisms. A single individual or species from this domain is called an archaeon...
shown to be associated with Iron Mountain belonged to the order Thermoplasmatales
Thermoplasmatales
In taxonomy, the Thermoplasmatales are an order of the Thermoplasmata. All are acidophiles, growing optimally at pH below 2. Picrophilus is currently the most acidophilic of all known organisms growing at a minimum pH of 0.06. Many of these organisms do not contain a cell wall, although this is...
(e.g., Ferroplasma
Ferroplasma
In taxonomy, Ferroplasma is a genus of the Ferroplasmaceae.The genus Ferroplasma consists solely of F. acidophilum, an acidophilic iron-oxidizing member of the Euryarchaeota. Unlike other members of the Thermoplasmata F.acidophilum is a mesophile with a temperature optimum of approximately 35°C, at...
acidarmanus).
Distribution
Examination of different sites in the mine using fluorescent probes specific to the ARMAN groups has revealed that they are always present in communities associated with acid mine drainageAcid mine drainage
Acid mine drainage , or acid rock drainage , refers to the outflow of acidic water from metal mines or coal mines. However, other areas where the earth has been disturbed may also contribute acid rock drainage to the environment...
(AMD), at Iron Mountain in northern California, that have pH < 1.5. They are usually found in low abundance (5–25%) in the community. More recently, closely related organisms have been detected in an acidic boreal mire or bog
Bog
A bog, quagmire or mire is a wetland that accumulates acidic peat, a deposit of dead plant material—often mosses or, in Arctic climates, lichens....
in Finland and another acid mine drainage site in extreme environments of Rio Tinto, southwestern Spain.
Cell structure and ecology
Using cryo-electron tomographyCryo-electron tomography
Cryo-electron tomography is a type of electron cryomicroscopy where tomography is used to obtain a 3D reconstruction of a sample from tilted 2D images at cryogenic temperatures....
an extensive 3D characterization of uncultivated ARMAN cells within mine biofilms has been done (Comolli et al. 2009). This has revealed that they are right at the cell size predicted to be the lower limit for life, 0.009 µm3 and 0.04 µm3 (NRC Steering group). They also found that despite their unusually small cell size it is common to find more than one type of virus
Virus
A virus is a small infectious agent that can replicate only inside the living cells of organisms. Viruses infect all types of organisms, from animals and plants to bacteria and archaea...
attached to the cells while in the biofilms. Furthermore, the cells contain on average ~92 ribosomes per cell, whereas the average E. coli cell grown in culture contains ~10,000 ribosomes. This suggests that for ARMAN cells a much more limited number of metabolites are present in a given cell. It raises questions about what the minimal requirements are for a living cell.
3D reconstructions of ARMAN cells in the environment has revealed that a small number of them attach to other Archaea of the order Thermoplasmatales
Thermoplasmatales
In taxonomy, the Thermoplasmatales are an order of the Thermoplasmata. All are acidophiles, growing optimally at pH below 2. Picrophilus is currently the most acidophilic of all known organisms growing at a minimum pH of 0.06. Many of these organisms do not contain a cell wall, although this is...
(Baker et al. 2010 ). The ARMAN cells appear to penetrate the cell wall to the cytoplasm of the Thermoplasmatales cells. The nature of this interaction hasn't been determined. It could be some sort of parasitic or symbiotic interaction. It's possible that ARMAN is getting some sort of metabolite that it is not able to produce on its own.
Genomics and proteomics
The genomes of three ARMAN groups have been sequenced at the DOE Joint Genome InstituteJoint Genome Institute
The U.S. Department of Energy Joint Genome Institute was created in 1997 to unite the expertise and resources in genome mapping, DNA sequencing, technology development, and information sciences pioneered at the DOE genome centers at Lawrence Berkeley National Laboratory , Lawrence Livermore...
during a 2006 Community Sequencing Program (CSP). These three genomes were successfully binned from the community genomic data using ESOM or Emergent Self-Organizing Map
Self-organizing map
A self-organizing map or self-organizing feature map is a type of artificial neural network that is trained using unsupervised learning to produce a low-dimensional , discretized representation of the input space of the training samples, called a map...
clustering of tetra-nucleotide DNA signatures (Dick et al. 2009).
The first draft of Candidatus Micrarchaeum acidiphilum ARMAN-2 is ~1 Mb (3 scaffolds, Baker et al.. PNAS 2010. The ARMAN-2 has recently been closed using 454 and Solexa sequencing of other biofilms to close the gaps and is being prepared for submission to NCBI. The genomes of ARMAN-4 and ARMAN-5 (roughly 1 Mb as well) have unusually average genes sizes, similar to those seen in endosymbiotic and parasitic bacteria. This may be signature of their inter-species interactions with other Archaea in nature (Baker et al. 2010 ). Furthermore, the branching of these groups near the Euryarchaea/Crenarchaea divide is reflected in them sharing many genetic aspects of both Crenarchaea and Euryarchaea. Specifically they have many genes that had previously only been identified in Crenarchaea. It is difficult to elucidate many of the commonly known metabolic pathways in ARMAN due to the unusually high number of unique genes that have been identified in their genomes.
A novel type of tRNA splicing endonuclease, involved in the processing of tRNA, has been discovered in ARMAN groups 1 and 2 (Fujishima et al. 2011). The enzyme consists of two duplicated catalytic units and one structural unit encoded on a single gene, representing a novel three-unit architecture.
External links
- ASM Small Things Considered blog article
- JGI Community Sequencing Program
- 2006 University of California Berkeley Press Release
- Dr. Luis Comolli's home page with several images of ARMAN cells
- 2010 Univ. of California Berkeley press release
- 2010 USA Today article about ARMAN
- 2010 MSNBC article about ARMAN

