U4 spliceosomal RNA
Encyclopedia
The U4 small nuclear Ribo-Nucleic Acid (U4 snRNA) is a non-coding RNA
component of the major or U2-dependent spliceosome
– a eukaryotic molecular machine
involved in the splicing of pre-messenger RNA (pre-mRNA). It forms a duplex with U6
, and with each splicing round, it is displaced from the U6 snRNA (and the spliceosome) in an ATP-dependent manner, allowing U6 to re-fold and create the active site
for splicing
catalysis. A recycling process involving protein Brr2 releases U4 from U6, while protein Prp24
re-anneals U4 and U6. The crystal structure
of a 5′ stem-loop
of U4 in complex with a binding protein has been solved.
, involved with the U6 snRNA in the di-snRNP, as well as involved with both the U6 snRNA and the U5 snRNA in the tri-snRNP. The different formats have been proposed to coincide with different temporal events in the activity of the penta-snRNP, or as intermediates in the step-wise model of spliceosome assembly and activity.
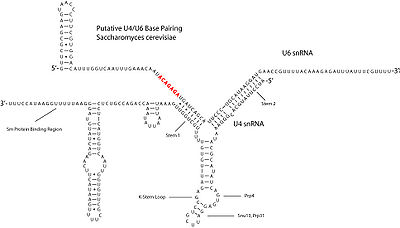
The U4 snRNA (and its likely analog snR14 in Yeast) has been shown not to participate directly in the specific catalytic activities of the splicing reaction, and is proposed instead to act as a regulator of the U6 snRNA. The U4 snRNA inhibits spliceosome activity during assembly by complementary base pairing between the U6 snRNA in two highly conserved stem regions. It is suggested that this base-pairing interaction prevents the U6 snRNA from assembling with the U2 snRNA into the conformation required for catalytic activity. If the U4 snRNA is degraded and thereby removed from the spliceosome, splicing is effectively halted. The U4 and U6 snRNAs are demonstratively required for splicing in vitro.
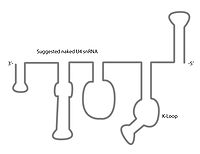
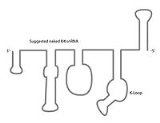
 The U4 snRNA secondary structure is suggested to alter depending on its interaction with the U6 snRNA. Several experiments involving X-ray crystallography
The U4 snRNA secondary structure is suggested to alter depending on its interaction with the U6 snRNA. Several experiments involving X-ray crystallography
, NMR
, and chemical modification RNA structure probing
indicate that U4 snRNA secondary structure contains several conserved motifs, which serve structural as well as intermediary roles in establishing interactions with other splicing components. The putative U4/U6 snRNA base pairing secondary structure shown in Figure 2., is conserved across a diverse set of organisms suggesting the splicing machinery's ancient origins. It has been shown previously that a highly conserved Kinked-loop participates in specific protein interactions.
Non-coding RNA
A non-coding RNA is a functional RNA molecule that is not translated into a protein. Less-frequently used synonyms are non-protein-coding RNA , non-messenger RNA and functional RNA . The term small RNA is often used for short bacterial ncRNAs...
component of the major or U2-dependent spliceosome
Spliceosome
A spliceosome is a complex of snRNA and protein subunits that removes introns from a transcribed pre-mRNA segment. This process is generally referred to as splicing.-Composition:...
– a eukaryotic molecular machine
Molecular machine
A molecular machine, or nanomachine, is any discrete number of molecular components that produce quasi-mechanical movements in response to specific stimuli . The expression is often more generally applied to molecules that simply mimic functions that occur at the macroscopic level...
involved in the splicing of pre-messenger RNA (pre-mRNA). It forms a duplex with U6
U6 spliceosomal RNA
U6 snRNA is the non-coding small nuclear RNA component of U6 snRNP , an RNA-protein complex that combines with other snRNPs, unmodified pre-mRNA, and various other proteins to assemble a spliceosome, a large RNA-protein molecular complex upon which splicing of pre-mRNA occurs...
, and with each splicing round, it is displaced from the U6 snRNA (and the spliceosome) in an ATP-dependent manner, allowing U6 to re-fold and create the active site
Active site
In biology the active site is part of an enzyme where substrates bind and undergo a chemical reaction. The majority of enzymes are proteins but RNA enzymes called ribozymes also exist. The active site of an enzyme is usually found in a cleft or pocket that is lined by amino acid residues that...
for splicing
RNA splicing
In molecular biology and genetics, splicing is a modification of an RNA after transcription, in which introns are removed and exons are joined. This is needed for the typical eukaryotic messenger RNA before it can be used to produce a correct protein through translation...
catalysis. A recycling process involving protein Brr2 releases U4 from U6, while protein Prp24
Prp24
Prp24 is a protein part of the pre-messenger RNA splicing process and aids the binding of U6 snRNA to U4 snRNA during the formation of spliceosomes. Found in eukaryotes from yeast to E. coli, fungi, and humans, Prp24 was initially discovered to be an important element of RNA splicing in 1989...
re-anneals U4 and U6. The crystal structure
Crystal structure
In mineralogy and crystallography, crystal structure is a unique arrangement of atoms or molecules in a crystalline liquid or solid. A crystal structure is composed of a pattern, a set of atoms arranged in a particular way, and a lattice exhibiting long-range order and symmetry...
of a 5′ stem-loop
Stem-loop
Stem-loop intramolecular base pairing is a pattern that can occur in single-stranded DNA or, more commonly, in RNA. The structure is also known as a hairpin or hairpin loop. It occurs when two regions of the same strand, usually complementary in nucleotide sequence when read in opposite directions,...
of U4 in complex with a binding protein has been solved.
Biological role
The U4 snRNA has been shown to exist in a number of different formats including: bound to proteins as a small nuclear Ribo-Nuclear Protein snRNPSnRNP
snRNPs , or small nuclear ribonucleoproteins, are RNA-protein complexes that combine with unmodified pre-mRNA and various other proteins to form a spliceosome, a large RNA-protein molecular complex upon which splicing of pre-mRNA occurs...
, involved with the U6 snRNA in the di-snRNP, as well as involved with both the U6 snRNA and the U5 snRNA in the tri-snRNP. The different formats have been proposed to coincide with different temporal events in the activity of the penta-snRNP, or as intermediates in the step-wise model of spliceosome assembly and activity.
The U4 snRNA (and its likely analog snR14 in Yeast) has been shown not to participate directly in the specific catalytic activities of the splicing reaction, and is proposed instead to act as a regulator of the U6 snRNA. The U4 snRNA inhibits spliceosome activity during assembly by complementary base pairing between the U6 snRNA in two highly conserved stem regions. It is suggested that this base-pairing interaction prevents the U6 snRNA from assembling with the U2 snRNA into the conformation required for catalytic activity. If the U4 snRNA is degraded and thereby removed from the spliceosome, splicing is effectively halted. The U4 and U6 snRNAs are demonstratively required for splicing in vitro.
Structure


X-ray crystallography
X-ray crystallography is a method of determining the arrangement of atoms within a crystal, in which a beam of X-rays strikes a crystal and causes the beam of light to spread into many specific directions. From the angles and intensities of these diffracted beams, a crystallographer can produce a...
, NMR
NMR
NMR may refer to:Applications of Nuclear Magnetic Resonance:* Nuclear magnetic resonance* NMR spectroscopy* Solid-state nuclear magnetic resonance* Protein nuclear magnetic resonance spectroscopy* Proton NMR* Carbon-13 NMR...
, and chemical modification RNA structure probing
RNA structure probing
Structure probing of nucleic acids is the process by which biochemical techniques are used to determine nucleic acid structure. This analysis can be used to define the patterns which can infer the molecular structure, experimental analysis of molecular structure and function, and further...
indicate that U4 snRNA secondary structure contains several conserved motifs, which serve structural as well as intermediary roles in establishing interactions with other splicing components. The putative U4/U6 snRNA base pairing secondary structure shown in Figure 2., is conserved across a diverse set of organisms suggesting the splicing machinery's ancient origins. It has been shown previously that a highly conserved Kinked-loop participates in specific protein interactions.

