
Pathogenomics
Encyclopedia
Pathogen
infections are among the leading causes of infirmity and mortality among humans and animals in the world. Until recently, it has been difficult to compile information to understand the generation of pathogen virulence factors as well as pathogen
behaviour in a host environment. The study of Pathogenomics attempts to utilize genomic and metagenomics
data gathered from high through-put technologies (e.g. sequencing
or DNA microarrays), to understand microbe diversity and interaction as well as host-microbe interactions involved in disease states. The bulk of pathogenomics research concerns itself with pathogens that affect human health; however, studies also exist for plant and animal infecting microbes.
, it was difficult and costly to obtain sequence information for any pathogen. In 1995, the first pathogen genome, that of Haemophilus influenza was sequenced by traditional Sanger methods. Sanger methods, however, were slow and costly. The emergence of second generation high through-put sequencing technologies have allowed for microbial sequence information to be obtained much more quickly and at a considerably lower cost. Largely thanks to second generation sequencing methods, hundreds of pathogen genomes have been sequenced since 1995. The emergence of second generation high through-put sequencing technologies have allowed for microbial sequence information to be obtained much more quickly and at a considerably lower cost. This influx of information is also due to the capacity of sequencing platforms to evaluate the sequences of many organisms in parallel.
With the sequences of many organisms available for analysis, scientists, through their investigations, began to challenge some of the earlier tenants of bacterial genome structure. Older paradigms of microbial genomics believed that only a few strains were sufficient to represent a specific bacterial species.
It was thought that bacterial genomes, like eukaryotes, were relatively stable. In 2001, however, the sequences of Escherichia coli
0157:H7 was obtained in a study by Perna and her colleges; the study showed that two members of the same bacterial species can differ as much as 30% in genomic content. It became evident that sequencing multiple strains for a species, rather than a few selectively chosen ones, was necessary to understand the diversity in a microbial species gene pool
. It was also increasingly important to understand how to account for these differences in genomic content across a species strains and how it may contribute to pathogenic behaviour or prevent the formation of pathogens.
More recently, the sequenced genomic data has been catalogued in databases and made publicly available online (there do also exist non-publicly available databases in the private sector
). The availability and influx of this information presses upon those who conduct pathogenomics research to come up with a way of drawing meaningful conclusions from this data. In addition, the availability of such data on the internet encourages global collaboration of labs.
l behaviour. More recently there have been increased efforts to sequence Eukarya genomes and more will be underway in the future. Regardless of this bias in reporting, many of the dynamic genomic events are similar across all the types of pathogen organisms.
Pathogenomics does not focus exclusively on understanding pathogen-host interaction. Insight of individual or cooperative pathogen behaviour provides knowledge into the development or inheritance of pathogen virulence factors. Through a deeper understanding of the small subunits that cause infection, it may be able possible to develop novel therapeutics that are efficient and cost effective.
technologies, it is possible to detect, compare and catalogue many of these dynamic genomic events. Particular interest is in understanding how genomic events lead to pathogen development and how these events may be interrupted to prevent it.
Gene loss or genome decay occurs when a gene is no longer used by the microbe or when a microbe attempts to adapt to a new ecological niche.
Sequencing efforts and microarray analysis have exposed a large number of pseudo genes in some bacterial pathogen species. Mycobacterium leprae
for example has been found to contain nearly as many pseudo genes as function genes. M. leprae is not the only microbe exhibiting such behaviour; in his article, Dr. Pallen reports similar properties from Yersinia pestis
(the plague pathogen) and also Salmonella enterica
. The inactivation of genes is typically associated with a change in the lifestyle of an organism, which can involve adapting to a new niche. The presence of extensive pseudogenes is contrary to another orthodox belief that all the genes in a bacterial genome are functional for some purpose
It is possible to detect the presence of pseudogenes and the marks of genome decay through whole-genome sequencing in combination with comparative genomics
Comparative genomics has helped to reveal that pathogens may favour losing genes in order to live in a host-associated niche and become endosymbionts. Sometimes the shedding of certain genes also renders a pathogen microbe harmless. The analysis of Listeria strains, for example, has shown that a reduced genome size has led to the generation of a non-pathogenic Listeria strain from a pathogenic precursor.
One of the key forces driving gene gain is thought to be horizontal (lateral) gene transfer (LGT).
It is of particular interest in microbial studies because these mobile genetic elements may introduce virulence factors into a new genome. An important comparative study conducted by Gill et al. in 2005 postulated that LGT may have been the cause for pathogen variations between Staphylococcus epidermidis
and Staphylococcus aureus
. There still, however, remains scepticism about the frequency of LGT, its identification, and its impact.
New and improved methodologies have been engaged, especially in the study of phylogenetics, to validate the presence and effect of LGT.
Gene gain and gene duplication events are balanced by gene loss, such that despite their dynamic nature, the genome of a bacterial species remains approximately the same size.
Mobile genetic insertion sequences can play a role in genome rearrangement activities. Pathogens that do not live in an isolated environment have been found to contain a large number of insertion sequence elements and various repetitive segments of DNA. The combination of these two genetic elements is thought help mediate homologous recombination
. There are pathogens, such as Burkholderia mallei
,
and Burkholderia pseudomallei
which have been shown to exhibit genome-wide rearrangements due to insertion sequences and repetitive DNA segments. At this time, no studies demonstrate genome-wide rearrangement events directly giving rise to pathogenic behaviour in a microbe. This does not mean it is not possible. Genome-wide rearrangements do, however, contribute to the plasticity of bacterial genome, which may prime the conditions for other factors to introduce, or lose, virulence factors.
Single nucleotide polymorphisms(SNPs) are also a genomic variable that adds to the diversity of pathogen strains. Current efforts attempt to catalogue the various SNPs in pathogen strains.
is a powerful methodology that has gained more applicability with the recent increased amount of sequence information. There are several examples of successful comparative genomics studies, among them the analysis of Listeria. and Escherichia coli. The most important topic comparative genomics, in a pathogenomic context, attempts to address the difference between pathogenic and non-pathogenic microbes. This inquiry, though, proves to be very difficult to analyze, since a single bacterial species can have many strains and the genomic content of each of these strains can vary.
The diversity within pathogen genomes makes it difficult to identify the total number of genes that are associated within all strains of a pathogen species. In fact, it has been thought that the total number of genes associated with a single pathogen species may be unlimited, although some groups are attempting to derive a more empirical value. For this reason it was necessary to introduce the concept of pan-genomes and core genomes. Pan-genome and core genome literature also tends to have a bias towards reporting on prokaryotic pathogen organisms. Caution may need to be exercised when extending the definition of a pan-genome or a core-genome to the other pathogen organisms; this is because there is no formal evidence of the properties of these pan-genomes. Here, it will be assumed that the definitions may in fact extend, since all pathogen organisms share in the same dynamic genomic events and rely upon variability within strains as a mechanism of survival and virulence.
A core genome is the set of genes found across all strains of a pathogen species. A pan-genome is the entire gene pool for that pathogen species, and includes genes that are not shared by all strains. Pan-genomes may be open or closed depending on whether comparative analysis of multiple strains reveals no new genes (closed) or many new genes (open) compared to the core genome for that pathogen species. In the open pan-genome, genes may be further characterized as dispensable or strain specific. Dispensable genes are those found in more than one strain, but not in all strains, of a pathogen species. Strain specific genes are those found only in one strain of a pathogen species. The differences in pan-genomes are reflections of the life style of the organism. For example, Streptococcus agalactiae
, which exists in diverse biological niches, has a broader pan-genome when compared with the more environmentally isolated Bacillus anthracis
. Comparative genomics
approaches are also being used to understand more about the pan-genome.
, and prophages. Pathogencity islands and their detection are the focous of several bioinformatics efforts involved in pathogenomics.
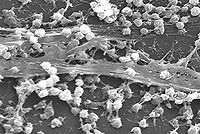 Microbe-host interactions tend to overshadow the consideration of microbe-microbe interactions. Microbe-microbe interactions though can lead to chronic states of infirmity that are difficult to understand and treat.
Microbe-host interactions tend to overshadow the consideration of microbe-microbe interactions. Microbe-microbe interactions though can lead to chronic states of infirmity that are difficult to understand and treat.
methods to form an expression profile of biofilm interacting bacteria. This expression profile may be used in subsequent analysis of other microbes to predict biofilm microbe behaviour, or to understand how to dismantle biofilm formation.
The diverse community within the gut has been heralded to be vital for human health. There a number of projects under way to better understand the ecosystems of the gut. The sequence of commensal Escherichia coli
strain SE11, for example, has already been determined from the faecal matter of a healthy human and promises to be the first of many studies. Through genomic analysis and also subsequent protein analysis a better understand of the beneficial properties of commensal flora will be investigated in hopes of understanding how to build a better therapeutic.
, are prominent demonstration of how over time a microbe, in this case Yersinia pestis
, evolves from a gastrointestinal pathogen to a very highly pathogenic microbe through dynamic genomic events. These flips between being pathogenic and non-pathogenic status and how they occurred with respect to ecological or environmental changes is important in novel therapeutic development to combat microbial infections.
. Bioinformatics
approaches are providing much of the power for rapidly mining, organising, analyzing, visualizing and annotating the data catalogued in databases.
Note that pathogenomics research could shed light for extensions of pathogens, or non-pathogens, that are not related to human, plant or animal health ; using microbes for bioremediation is one example. There is some but very little dialogue, however, concerning these extensions to pathogens and their relation to pathogenomics. It would be more suitable to categorize pathogen
/non-pathogen applications that are unrelated to infection under the more general category of microbe genomics. Some general reviews speak extensively both about pathogen related and non-related applications in the same article.
In the advent of new technology, it is easy to forget some of the basic things that prevent pathogen infections from starting and spreading. While there of course exists more deadly and difficult to handle pathogen infections, there do also exist less dangerous ones. Historically, human health greatly improved with more emphasis on healthy life styles including better hygiene
practices and access to clean recourses of water and nutrition. While pathogenomics can help provide insights into treatment and detection of some less potent pathogens, it must be kept in mind that there are many pathogens and only so much funding.
variation cannot match pathogen variation. Reverse vaccinology
is a novel approach that may develop vaccines to combat pathogens more effectively. Reverse Vaccinology has already been successfully applied to Neisseria meningitides, Streptococcus pneumoniae
and Chlamydia
spp. Reverse Vaccinology applies not only to strain specific vaccines, but also the development of pan-genome vaccines. Lastly, comparative vaccinology attempts to compare the differences between pathogen and non-pathogen variants of a microbe to filter genes that are unique to the pathogen version. There are several vaccines developed through reverse Vaccinology that are currently in clinical trials.
shed light on the possibility of the bioterrorism as being more of a real than imagined threat. Bioterrorism
was anticipated in the Iraq war, with soldiers even being inoculated for an small pox attack. Using technologies and insight gained from reconstruction the Spanish influenza, it may be possible to prevent future deadly planted outbreaks of disease. There is a strong ethical concern however, as to whether the resurrection of old viruses is necessary and whether it is actually more harm than good.
Pathogen
A pathogen gignomai "I give birth to") or infectious agent — colloquially, a germ — is a microbe or microorganism such as a virus, bacterium, prion, or fungus that causes disease in its animal or plant host...
infections are among the leading causes of infirmity and mortality among humans and animals in the world. Until recently, it has been difficult to compile information to understand the generation of pathogen virulence factors as well as pathogen
Pathogen
A pathogen gignomai "I give birth to") or infectious agent — colloquially, a germ — is a microbe or microorganism such as a virus, bacterium, prion, or fungus that causes disease in its animal or plant host...
behaviour in a host environment. The study of Pathogenomics attempts to utilize genomic and metagenomics
Metagenomics
Metagenomics is the study of metagenomes, genetic material recovered directly from environmental samples. The broad field may also be referred to as environmental genomics, ecogenomics or community genomics. Traditional microbiology and microbial genome sequencing rely upon cultivated clonal cultures...
data gathered from high through-put technologies (e.g. sequencing
Sequencing
In genetics and biochemistry, sequencing means to determine the primary structure of an unbranched biopolymer...
or DNA microarrays), to understand microbe diversity and interaction as well as host-microbe interactions involved in disease states. The bulk of pathogenomics research concerns itself with pathogens that affect human health; however, studies also exist for plant and animal infecting microbes.
History
In the early investigation of microbial genomicsGenomics
Genomics is a discipline in genetics concerning the study of the genomes of organisms. The field includes intensive efforts to determine the entire DNA sequence of organisms and fine-scale genetic mapping efforts. The field also includes studies of intragenomic phenomena such as heterosis,...
, it was difficult and costly to obtain sequence information for any pathogen. In 1995, the first pathogen genome, that of Haemophilus influenza was sequenced by traditional Sanger methods. Sanger methods, however, were slow and costly. The emergence of second generation high through-put sequencing technologies have allowed for microbial sequence information to be obtained much more quickly and at a considerably lower cost. Largely thanks to second generation sequencing methods, hundreds of pathogen genomes have been sequenced since 1995. The emergence of second generation high through-put sequencing technologies have allowed for microbial sequence information to be obtained much more quickly and at a considerably lower cost. This influx of information is also due to the capacity of sequencing platforms to evaluate the sequences of many organisms in parallel.
With the sequences of many organisms available for analysis, scientists, through their investigations, began to challenge some of the earlier tenants of bacterial genome structure. Older paradigms of microbial genomics believed that only a few strains were sufficient to represent a specific bacterial species.
It was thought that bacterial genomes, like eukaryotes, were relatively stable. In 2001, however, the sequences of Escherichia coli
Escherichia coli
Escherichia coli is a Gram-negative, rod-shaped bacterium that is commonly found in the lower intestine of warm-blooded organisms . Most E. coli strains are harmless, but some serotypes can cause serious food poisoning in humans, and are occasionally responsible for product recalls...
0157:H7 was obtained in a study by Perna and her colleges; the study showed that two members of the same bacterial species can differ as much as 30% in genomic content. It became evident that sequencing multiple strains for a species, rather than a few selectively chosen ones, was necessary to understand the diversity in a microbial species gene pool
Gene pool
In population genetics, a gene pool is the complete set of unique alleles in a species or population.- Description :A large gene pool indicates extensive genetic diversity, which is associated with robust populations that can survive bouts of intense selection...
. It was also increasingly important to understand how to account for these differences in genomic content across a species strains and how it may contribute to pathogenic behaviour or prevent the formation of pathogens.
More recently, the sequenced genomic data has been catalogued in databases and made publicly available online (there do also exist non-publicly available databases in the private sector
Private sector
In economics, the private sector is that part of the economy, sometimes referred to as the citizen sector, which is run by private individuals or groups, usually as a means of enterprise for profit, and is not controlled by the state...
). The availability and influx of this information presses upon those who conduct pathogenomics research to come up with a way of drawing meaningful conclusions from this data. In addition, the availability of such data on the internet encourages global collaboration of labs.
Microbe Analysis
Pathogens may be prokaryotic (archaea or bacteria), single-celled Eukarya or viruses. Prokaryotic genomes have typically been easier to sequence due to smaller genome size compared to Eukarya. Due to this, there is a bias in reporting pathogenic bacteriaPathogenic bacteria
Pathogenic bacteria are bacteria that cause bacterial infection. This article deals with human pathogenic bacteria.Although the vast majority of bacteria are harmless or beneficial, quite a few bacteria are pathogenic...
l behaviour. More recently there have been increased efforts to sequence Eukarya genomes and more will be underway in the future. Regardless of this bias in reporting, many of the dynamic genomic events are similar across all the types of pathogen organisms.
Pathogenomics does not focus exclusively on understanding pathogen-host interaction. Insight of individual or cooperative pathogen behaviour provides knowledge into the development or inheritance of pathogen virulence factors. Through a deeper understanding of the small subunits that cause infection, it may be able possible to develop novel therapeutics that are efficient and cost effective.
Analyzing individual microbes
Dynamic genomes with high plasticity are necessary to allow pathogens, especially bacteria, to survive in changing environments. With the assistance of high throughput sequencing methods and in silicoIn silico
In silico is an expression used to mean "performed on computer or via computer simulation." The phrase was coined in 1989 as an analogy to the Latin phrases in vivo and in vitro which are commonly used in biology and refer to experiments done in living organisms and outside of living organisms,...
technologies, it is possible to detect, compare and catalogue many of these dynamic genomic events. Particular interest is in understanding how genomic events lead to pathogen development and how these events may be interrupted to prevent it.
Causes of Genomic Diversity
Three forces act in shaping the pathogen genome: gene gain, gene loss, and genome rearrangement. The knowledge and detection of these genomic dynamic events are necessary in the construction of useful therapeutic tools to combat pathogens.Gene Loss / Genome Decay
Gene loss or genome decay occurs when a gene is no longer used by the microbe or when a microbe attempts to adapt to a new ecological niche.
Sequencing efforts and microarray analysis have exposed a large number of pseudo genes in some bacterial pathogen species. Mycobacterium leprae
Mycobacterium leprae
Mycobacterium leprae, also known as Hansen’s coccus spirilly, mostly found in warm tropical countries, is a bacterium that causes leprosy . It is an intracellular, pleomorphic, acid-fast bacterium. M. leprae is an aerobic bacillus surrounded by the characteristic waxy coating unique to mycobacteria...
for example has been found to contain nearly as many pseudo genes as function genes. M. leprae is not the only microbe exhibiting such behaviour; in his article, Dr. Pallen reports similar properties from Yersinia pestis
Yersinia pestis
Yersinia pestis is a Gram-negative rod-shaped bacterium. It is a facultative anaerobe that can infect humans and other animals....
(the plague pathogen) and also Salmonella enterica
Salmonella enterica
Salmonella enterica is a rod-shaped flagellated, facultative anaerobic, Gram-negative bacterium, and a member of the genus Salmonella.- Epidemiology :...
. The inactivation of genes is typically associated with a change in the lifestyle of an organism, which can involve adapting to a new niche. The presence of extensive pseudogenes is contrary to another orthodox belief that all the genes in a bacterial genome are functional for some purpose
It is possible to detect the presence of pseudogenes and the marks of genome decay through whole-genome sequencing in combination with comparative genomics
Comparative genomics
Comparative genomics is the study of the relationship of genome structure and function across different biological species or strains. Comparative genomics is an attempt to take advantage of the information provided by the signatures of selection to understand the function and evolutionary...
Comparative genomics has helped to reveal that pathogens may favour losing genes in order to live in a host-associated niche and become endosymbionts. Sometimes the shedding of certain genes also renders a pathogen microbe harmless. The analysis of Listeria strains, for example, has shown that a reduced genome size has led to the generation of a non-pathogenic Listeria strain from a pathogenic precursor.
Gene Gain/ Gene Duplication
One of the key forces driving gene gain is thought to be horizontal (lateral) gene transfer (LGT).
It is of particular interest in microbial studies because these mobile genetic elements may introduce virulence factors into a new genome. An important comparative study conducted by Gill et al. in 2005 postulated that LGT may have been the cause for pathogen variations between Staphylococcus epidermidis
Staphylococcus epidermidis
Staphylococcus epidermidis is one of thirty-three known species belonging to the genus Staphylococcus. It is part of human skin flora, and consequently part of human flora. It can also be found in the mucous membranes and in animals. Due to contamination, it is probably the most common species...
and Staphylococcus aureus
Staphylococcus aureus
Staphylococcus aureus is a facultative anaerobic Gram-positive coccal bacterium. It is frequently found as part of the normal skin flora on the skin and nasal passages. It is estimated that 20% of the human population are long-term carriers of S. aureus. S. aureus is the most common species of...
. There still, however, remains scepticism about the frequency of LGT, its identification, and its impact.
New and improved methodologies have been engaged, especially in the study of phylogenetics, to validate the presence and effect of LGT.
Gene gain and gene duplication events are balanced by gene loss, such that despite their dynamic nature, the genome of a bacterial species remains approximately the same size.
Genome Rearrangement
Mobile genetic insertion sequences can play a role in genome rearrangement activities. Pathogens that do not live in an isolated environment have been found to contain a large number of insertion sequence elements and various repetitive segments of DNA. The combination of these two genetic elements is thought help mediate homologous recombination
Homologous recombination
Homologous recombination is a type of genetic recombination in which nucleotide sequences are exchanged between two similar or identical molecules of DNA. It is most widely used by cells to accurately repair harmful breaks that occur on both strands of DNA, known as double-strand breaks...
. There are pathogens, such as Burkholderia mallei
Burkholderia mallei
Burkholderia mallei is a gram-negative bipolar aerobic bacterium, a Burkholderia-genus human and animal pathogen causing Glanders; the Latin name of this disease gave name to the causative agent species...
,
and Burkholderia pseudomallei
Burkholderia pseudomallei
Burkholderia pseudomallei is a Gram-negative, bipolar, aerobic, motile rod-shaped bacterium. It infects humans and animals and causes the disease melioidosis. It is also capable of infecting plants....
which have been shown to exhibit genome-wide rearrangements due to insertion sequences and repetitive DNA segments. At this time, no studies demonstrate genome-wide rearrangement events directly giving rise to pathogenic behaviour in a microbe. This does not mean it is not possible. Genome-wide rearrangements do, however, contribute to the plasticity of bacterial genome, which may prime the conditions for other factors to introduce, or lose, virulence factors.
Single Nucleotide Polymorphism
Single nucleotide polymorphisms(SNPs) are also a genomic variable that adds to the diversity of pathogen strains. Current efforts attempt to catalogue the various SNPs in pathogen strains.
Analysis of Genomic Diversity
There is a need to analyze more than a single genome sequence of a pathogen species to understand pathogen mechanisms. Comparative genomicsComparative genomics
Comparative genomics is the study of the relationship of genome structure and function across different biological species or strains. Comparative genomics is an attempt to take advantage of the information provided by the signatures of selection to understand the function and evolutionary...
is a powerful methodology that has gained more applicability with the recent increased amount of sequence information. There are several examples of successful comparative genomics studies, among them the analysis of Listeria. and Escherichia coli. The most important topic comparative genomics, in a pathogenomic context, attempts to address the difference between pathogenic and non-pathogenic microbes. This inquiry, though, proves to be very difficult to analyze, since a single bacterial species can have many strains and the genomic content of each of these strains can vary.
Pan-genomes and core genomes
The diversity within pathogen genomes makes it difficult to identify the total number of genes that are associated within all strains of a pathogen species. In fact, it has been thought that the total number of genes associated with a single pathogen species may be unlimited, although some groups are attempting to derive a more empirical value. For this reason it was necessary to introduce the concept of pan-genomes and core genomes. Pan-genome and core genome literature also tends to have a bias towards reporting on prokaryotic pathogen organisms. Caution may need to be exercised when extending the definition of a pan-genome or a core-genome to the other pathogen organisms; this is because there is no formal evidence of the properties of these pan-genomes. Here, it will be assumed that the definitions may in fact extend, since all pathogen organisms share in the same dynamic genomic events and rely upon variability within strains as a mechanism of survival and virulence.
A core genome is the set of genes found across all strains of a pathogen species. A pan-genome is the entire gene pool for that pathogen species, and includes genes that are not shared by all strains. Pan-genomes may be open or closed depending on whether comparative analysis of multiple strains reveals no new genes (closed) or many new genes (open) compared to the core genome for that pathogen species. In the open pan-genome, genes may be further characterized as dispensable or strain specific. Dispensable genes are those found in more than one strain, but not in all strains, of a pathogen species. Strain specific genes are those found only in one strain of a pathogen species. The differences in pan-genomes are reflections of the life style of the organism. For example, Streptococcus agalactiae
Streptococcus agalactiae
Streptococcus agalactiae is a beta-hemolytic Gram-positive streptococcus.- Identification :The CAMP test is an important test for identification...
, which exists in diverse biological niches, has a broader pan-genome when compared with the more environmentally isolated Bacillus anthracis
Bacillus anthracis
Bacillus anthracis is the pathogen of the Anthrax acute disease. It is a Gram-positive, spore-forming, rod-shaped bacterium, with a width of 1-1.2µm and a length of 3-5µm. It can be grown in an ordinary nutrient medium under aerobic or anaerobic conditions.It is one of few bacteria known to...
. Comparative genomics
Comparative genomics
Comparative genomics is the study of the relationship of genome structure and function across different biological species or strains. Comparative genomics is an attempt to take advantage of the information provided by the signatures of selection to understand the function and evolutionary...
approaches are also being used to understand more about the pan-genome.
Mobile Genetic Elements that Encode Virulence factors
Three genetic elements of human-affecting pathogens contribute to the transfer of virulence factors: plasmids, pathogenicity islandPathogenicity island
Pathogenicity islands are a distinct class of genomic islands acquired by microorganisms through horizontal gene transfer. They are incorporated in the genome of pathogenic organisms but are usually absent from those non-pathogenic organisms of the same or closely related species...
, and prophages. Pathogencity islands and their detection are the focous of several bioinformatics efforts involved in pathogenomics.
Analyzing Microbe-Microbe Interactions

Bioflims
Biofilms are an example of microbe-microbe interactions and are thought to be associated with up to 80% of human infections. Recently it has been shown that there are specific genes and cell surface proteins involved in the formation of biofilm. These genes and also surface proteins may be characterized through in silicoIn silico
In silico is an expression used to mean "performed on computer or via computer simulation." The phrase was coined in 1989 as an analogy to the Latin phrases in vivo and in vitro which are commonly used in biology and refer to experiments done in living organisms and outside of living organisms,...
methods to form an expression profile of biofilm interacting bacteria. This expression profile may be used in subsequent analysis of other microbes to predict biofilm microbe behaviour, or to understand how to dismantle biofilm formation.
Host Microbe Analysis
A microbe may be influenced by hosts to either adapt to their new environment or learn to evade it. An insight into these behaviours will provide beneficial insight for potential therapeutics. The most detailed outline of host-microbe interaction initiatives is outlined by the Pathogenomics European Research Agenda. Its report emphasises the following features:- Microarray analysis of host and microbe gene expression during infection. This is important for identifying the expression of virulence factors that allow a pathogen to survive a host's defence mechanism. Pathogens tend to undergo an assortment of changed in order to subvert and hosts immune system, in some case favouring a hyper variable genome state. The genomic expression studies will be complimented with protein-protein interaction networks studies.
- Using RNA interference (RNAi) to identify host cell functions in response to infections. Infection depends on the balance between the characteristics of the host cell and the pathogen cell. In some cases, there can be an overactive host response to infection, such as in meningitis, which can overwhelm the host's body. Using RNA, it will be possible to more clearly identify how a host cell defends itself during times of acute or chronic infection. This has also been applied successfully is Drosophila.
- Not all microbe interactions in host environment are malicious. Commensal flora, which exists in various environments in animals and humans may actually help combating microbial infections. The human flora, such as the gut for example, is home to a myriad of microbes.
The diverse community within the gut has been heralded to be vital for human health. There a number of projects under way to better understand the ecosystems of the gut. The sequence of commensal Escherichia coli
Escherichia coli
Escherichia coli is a Gram-negative, rod-shaped bacterium that is commonly found in the lower intestine of warm-blooded organisms . Most E. coli strains are harmless, but some serotypes can cause serious food poisoning in humans, and are occasionally responsible for product recalls...
strain SE11, for example, has already been determined from the faecal matter of a healthy human and promises to be the first of many studies. Through genomic analysis and also subsequent protein analysis a better understand of the beneficial properties of commensal flora will be investigated in hopes of understanding how to build a better therapeutic.
Eco-evo perspective
The "eco-evo" perspective on pathogen-host interactions emphasizes the influences ecology and the environment on pathogen evolution. The dynamic genomic factors such as gene loss, gene gain and genome rearrangement are all strongly influenced by changes in the ecological niche that a particular microbial strain resides in. Microbes may switch from being pathogenic and non-pathogenic due to changing environments. Studies of the plague, Yersinia pestisYersinia pestis
Yersinia pestis is a Gram-negative rod-shaped bacterium. It is a facultative anaerobe that can infect humans and other animals....
, are prominent demonstration of how over time a microbe, in this case Yersinia pestis
Yersinia pestis
Yersinia pestis is a Gram-negative rod-shaped bacterium. It is a facultative anaerobe that can infect humans and other animals....
, evolves from a gastrointestinal pathogen to a very highly pathogenic microbe through dynamic genomic events. These flips between being pathogenic and non-pathogenic status and how they occurred with respect to ecological or environmental changes is important in novel therapeutic development to combat microbial infections.
Applications
Many of the future challenges of pathogenomics begin with handling and making sense of the large influx of data that now is available to the research community. Mining the data for useful information proves to be applicable to many facets of epidemiologyEpidemiology
Epidemiology is the study of health-event, health-characteristic, or health-determinant patterns in a population. It is the cornerstone method of public health research, and helps inform policy decisions and evidence-based medicine by identifying risk factors for disease and targets for preventive...
. Bioinformatics
Bioinformatics
Bioinformatics is the application of computer science and information technology to the field of biology and medicine. Bioinformatics deals with algorithms, databases and information systems, web technologies, artificial intelligence and soft computing, information and computation theory, software...
approaches are providing much of the power for rapidly mining, organising, analyzing, visualizing and annotating the data catalogued in databases.
Note that pathogenomics research could shed light for extensions of pathogens, or non-pathogens, that are not related to human, plant or animal health ; using microbes for bioremediation is one example. There is some but very little dialogue, however, concerning these extensions to pathogens and their relation to pathogenomics. It would be more suitable to categorize pathogen
Pathogen
A pathogen gignomai "I give birth to") or infectious agent — colloquially, a germ — is a microbe or microorganism such as a virus, bacterium, prion, or fungus that causes disease in its animal or plant host...
/non-pathogen applications that are unrelated to infection under the more general category of microbe genomics. Some general reviews speak extensively both about pathogen related and non-related applications in the same article.
In the advent of new technology, it is easy to forget some of the basic things that prevent pathogen infections from starting and spreading. While there of course exists more deadly and difficult to handle pathogen infections, there do also exist less dangerous ones. Historically, human health greatly improved with more emphasis on healthy life styles including better hygiene
Hygiene
Hygiene refers to the set of practices perceived by a community to be associated with the preservation of health and healthy living. While in modern medical sciences there is a set of standards of hygiene recommended for different situations, what is considered hygienic or not can vary between...
practices and access to clean recourses of water and nutrition. While pathogenomics can help provide insights into treatment and detection of some less potent pathogens, it must be kept in mind that there are many pathogens and only so much funding.
Reverse Vaccinology
The variability of genomes can make the developed of a vaccine very difficult, and antigenAntigen
An antigen is a foreign molecule that, when introduced into the body, triggers the production of an antibody by the immune system. The immune system will then kill or neutralize the antigen that is recognized as a foreign and potentially harmful invader. These invaders can be molecules such as...
variation cannot match pathogen variation. Reverse vaccinology
Reverse vaccinology
Reverse vaccinology is an improvement on vaccinology, pioneered by Rino Rappuoli and first used against meningococcus. Since then, it has been used on several other organisms.-Computational approach:...
is a novel approach that may develop vaccines to combat pathogens more effectively. Reverse Vaccinology has already been successfully applied to Neisseria meningitides, Streptococcus pneumoniae
Streptococcus pneumoniae
Streptococcus pneumoniae, or pneumococcus, is Gram-positive, alpha-hemolytic, aerotolerant anaerobic member of the genus Streptococcus. A significant human pathogenic bacterium, S...
and Chlamydia
Chlamydia (bacterium)
Chlamydia is a genus of bacteria that are obligate intracellular parasites. Chlamydia infections are the most common bacterial sexually transmitted infections in humans and are the leading cause of infectious blindness worldwide....
spp. Reverse Vaccinology applies not only to strain specific vaccines, but also the development of pan-genome vaccines. Lastly, comparative vaccinology attempts to compare the differences between pathogen and non-pathogen variants of a microbe to filter genes that are unique to the pathogen version. There are several vaccines developed through reverse Vaccinology that are currently in clinical trials.
Countering Bioterrorism
In 2005 the sequence of the 1918 Spanish influenza was completed. Accompanied with phylogenetic analysis, it was possible to supply a detailed account of the virus’ evolution and behaviour, in particular it’s adaptation to humans. Following the sequencing of the Spanish influenza, the pathogen was also reconstructed. When inserted into mice, the pathogen proved to be incredibly deadly. The 2001 anthrax attacks2001 anthrax attacks
The 2001 anthrax attacks in the United States, also known as Amerithrax from its Federal Bureau of Investigation case name, occurred over the course of several weeks beginning on Tuesday, September 18, 2001, one week after the September 11 attacks. Letters containing anthrax spores were mailed to...
shed light on the possibility of the bioterrorism as being more of a real than imagined threat. Bioterrorism
Bioterrorism
Bioterrorism is terrorism involving the intentional release or dissemination of biological agents. These agents are bacteria, viruses, or toxins, and may be in a naturally occurring or a human-modified form. For the use of this method in warfare, see biological warfare.-Definition:According to the...
was anticipated in the Iraq war, with soldiers even being inoculated for an small pox attack. Using technologies and insight gained from reconstruction the Spanish influenza, it may be possible to prevent future deadly planted outbreaks of disease. There is a strong ethical concern however, as to whether the resurrection of old viruses is necessary and whether it is actually more harm than good.

