
Microfluidic Sanger Sequencing
Encyclopedia
The completion of the Human Genome Project
has been a cornerstone in the advancement of biological studies. The outcomes of obtaining a complete reference map (including the sequence) of the human genome have ushered in the post-genome era of studies. Genomics
will (if it hasn’t already) revolutionize medicine, forensics, molecular biology, biotechnology, and many other related and even unrelated disciplines in the future.
Sequencing of DNA
has largely been based on dideoxy chain termination
developed by Sanger
et al. However, the ability of the Human Genome Project in obtaining the full human genomic sequence meant that modifications were required to be made to this method. In particular, the incorporation of technological innovation, making sequencing automated and high-throughput, made this decade-long worldwide effort successful .
Briefly, in its modern inception, high-throughput genome sequencing (also referred to as Whole Genome Shot-gun Sequencing
) involves fragmenting the genome into small single-stranded pieces, followed by amplification of the fragments by Polymerase Chain Reaction (PCR). Adopting the Sanger method, each DNA fragment is irreversibly terminated with the incorporation of a fluorescently labeled dideoxy chain-terminating nucleotide, thereby producing a DNA “ladder” of fragments that each differ in length by one base and bear a base-specific fluorescent label at the terminal base. Amplified base ladders are then separated by Capillary Array Electrophoresis (CAE) with automated, in situ “finish-line” detection of the fluorescently labeled ssDNA fragments, which provides an ordered sequence of the fragments. These sequence reads are then computer assembled into overlapping or contiguous sequences (termed "contigs") which resemble the full genomic sequence once fully assembled.
Rapid technological developments have now emerged as a result of the Human Genome Project. In particular Massively Parallel Sequencing (MPS) approaches such as those now in wide commercial use (Illumina/Solexa, Roche/454 Pyrosequencing, and ABI SOLiD) are proving to be attractive tools for sequencing. Typically, MPS methods can only obtain short read lengths (35bp with Illumina platforms to a maximum of 200-300bp by 454 Pyrosequencing).
Sanger Methods on the other hand achieve read lengths of approximately 800bp (typically 500-600bp with non-enriched DNA). The longer read lengths in Sanger methods display significant advantages over MPS tools especially in terms of sequencing repetitive regions of the genome. A challenge of short-read sequence data is particularly an issue in sequencing new genomes (de novo) and in sequencing highly rearranged genome segments, typically those seen of cancer genomes or in regions of chromosomes that exhibit structural variation.
application for DNA sequencing, in which the Sanger sequencing steps (thermal cycling, sample purification, and capillary electrophoresis) are integrated on a wafer-scale chip using nanoliter-scale sample volumes. This technology generates long and accurate sequence reads, while obviating many of the significant shortcomings of the conventional Sanger method (e.g. high consumption of expensive reagents, reliance on expensive equipment, personnel-intensive manipulations, etc.) by integrating and automating the Sanger sequencing steps.
(SNP) detection, single-strand conformation polymorphism
(SSCP) hetroduplex analysis, and short tandem repeat
(STR) analysis. Resolving DNA fragments according to differences in size and/or conformation is the most critical step in studying these features of the genome.
The sequencing chip has a four-layer construction, consisting of three 100-mm-diameter glass wafers (on which device elements are microfabricated) and a polydimethylsiloxane (PDMS) membrane. Reaction chambers and capillary electrophoresis channels are etched between the top two glass wafers, which are thermally bonded. Three-dimensional channel interconnections and microvalves are formed by the PDMS and bottom manifold glass wafer.
The device consists of three functional units, each corresponding to the Sanger sequencing steps. The Thermal Cycling (TC) unit is a 250-nanoliter reaction chamber with integrated resistive temperature detector, microvalves, and a surface heater. Movement of reagent between the top all-glass layer and the lower glass-PDMS layer occurs through 500-μm-diameter via-holes. After thermal-cycling, the reaction mixture undergoes purification in the capture/purification chamber, and then is injected into the capillary electrophoresis (CE) chamber. The CE unit consists of a 30-cm capillary which is folded into a compact switchback pattern via 65-μm-wide turns.
In the TC reaction chamber, dye-terminator sequencing reagent, template DNA, and primers are loaded into the TC chamber and thermal-cycled for 35 cycles ( at 95°C for 12 seconds and at 60°C for 55 seconds).
The charged reaction mixture (containing extension fragments, template DNA, and excess sequencing reagent) is conducted through a capture/purification chamber at 30°C via a 33-Volts/cm electric field applied between capture outlet and inlet ports. The capture gel through which the sample is driven, consists of 40 μM of oligonucleotide (complementary to the primers) covalently bound to a polyacrylamide matrix. Extension fragments are immobilized by the gel matrix, and excess primer, template, free nucleotides, and salts are eluted through the capture waste port. The capture gel is heated to 67-75°C to release extension fragments.
Extension fragments are injected into the CE chamber where they are electrophoresed through a 125-167-V/cm field.
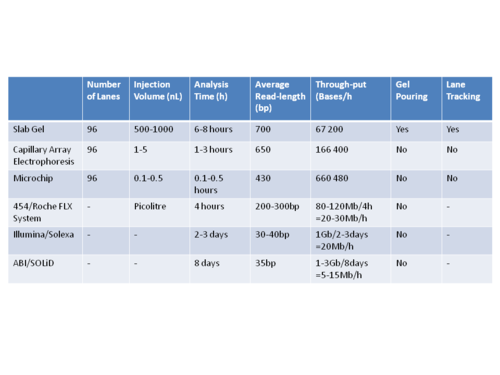 The ultimate goal of high-throughput sequencing is to develop systems that are low-cost, and extremely efficient at obtaining extended (longer) read lengths. Longer read lengths of each single electrophoretic separation, substantially reduces the cost associated with de novo DNA sequencing and the number of templates needed to sequence DNA contigs at a given redundancy. Microfluidics may allow for faster, cheaper and easier sequence assembly.
The ultimate goal of high-throughput sequencing is to develop systems that are low-cost, and extremely efficient at obtaining extended (longer) read lengths. Longer read lengths of each single electrophoretic separation, substantially reduces the cost associated with de novo DNA sequencing and the number of templates needed to sequence DNA contigs at a given redundancy. Microfluidics may allow for faster, cheaper and easier sequence assembly.
Help:http://en.wikipedia.org/wiki/DNA_sequencing#Chain-termination_methods
http://nano.cancer.gov/news_center/nanotech_news_2006-05-30a.asp
http://nano.cancer.gov/news_center/monthly_feature_2005_aug.asp
http://www.genomeweb.com/sequencing/mbi-says-new-tool-automates-sanger-sample-prep-cuts-reagent-and-labor-costs
Commercial Websites:
Illumina/Solexa Website
Roche 454 Life Sciences Website
ABI SOLiD website
Human Genome Project
The Human Genome Project is an international scientific research project with a primary goal of determining the sequence of chemical base pairs which make up DNA, and of identifying and mapping the approximately 20,000–25,000 genes of the human genome from both a physical and functional...
has been a cornerstone in the advancement of biological studies. The outcomes of obtaining a complete reference map (including the sequence) of the human genome have ushered in the post-genome era of studies. Genomics
Genomics
Genomics is a discipline in genetics concerning the study of the genomes of organisms. The field includes intensive efforts to determine the entire DNA sequence of organisms and fine-scale genetic mapping efforts. The field also includes studies of intragenomic phenomena such as heterosis,...
will (if it hasn’t already) revolutionize medicine, forensics, molecular biology, biotechnology, and many other related and even unrelated disciplines in the future.
Sequencing of DNA
DNA
Deoxyribonucleic acid is a nucleic acid that contains the genetic instructions used in the development and functioning of all known living organisms . The DNA segments that carry this genetic information are called genes, but other DNA sequences have structural purposes, or are involved in...
has largely been based on dideoxy chain termination
DNA sequencing
DNA sequencing includes several methods and technologies that are used for determining the order of the nucleotide bases—adenine, guanine, cytosine, and thymine—in a molecule of DNA....
developed by Sanger
Frederick Sanger
Frederick Sanger, OM, CH, CBE, FRS is an English biochemist and a two-time Nobel laureate in chemistry, the only person to have been so. In 1958 he was awarded a Nobel prize in chemistry "for his work on the structure of proteins, especially that of insulin"...
et al. However, the ability of the Human Genome Project in obtaining the full human genomic sequence meant that modifications were required to be made to this method. In particular, the incorporation of technological innovation, making sequencing automated and high-throughput, made this decade-long worldwide effort successful .
Briefly, in its modern inception, high-throughput genome sequencing (also referred to as Whole Genome Shot-gun Sequencing
Shotgun sequencing
In genetics, shotgun sequencing, also known as shotgun cloning, is a method used for sequencing long DNA strands. It is named by analogy with the rapidly-expanding, quasi-random firing pattern of a shotgun....
) involves fragmenting the genome into small single-stranded pieces, followed by amplification of the fragments by Polymerase Chain Reaction (PCR). Adopting the Sanger method, each DNA fragment is irreversibly terminated with the incorporation of a fluorescently labeled dideoxy chain-terminating nucleotide, thereby producing a DNA “ladder” of fragments that each differ in length by one base and bear a base-specific fluorescent label at the terminal base. Amplified base ladders are then separated by Capillary Array Electrophoresis (CAE) with automated, in situ “finish-line” detection of the fluorescently labeled ssDNA fragments, which provides an ordered sequence of the fragments. These sequence reads are then computer assembled into overlapping or contiguous sequences (termed "contigs") which resemble the full genomic sequence once fully assembled.
Rapid technological developments have now emerged as a result of the Human Genome Project. In particular Massively Parallel Sequencing (MPS) approaches such as those now in wide commercial use (Illumina/Solexa, Roche/454 Pyrosequencing, and ABI SOLiD) are proving to be attractive tools for sequencing. Typically, MPS methods can only obtain short read lengths (35bp with Illumina platforms to a maximum of 200-300bp by 454 Pyrosequencing).
Sanger Methods on the other hand achieve read lengths of approximately 800bp (typically 500-600bp with non-enriched DNA). The longer read lengths in Sanger methods display significant advantages over MPS tools especially in terms of sequencing repetitive regions of the genome. A challenge of short-read sequence data is particularly an issue in sequencing new genomes (de novo) and in sequencing highly rearranged genome segments, typically those seen of cancer genomes or in regions of chromosomes that exhibit structural variation.
Microfluidic Sanger Sequencing
Microfluidic Sanger sequencing is a lab-on-a-chipLab-on-a-chip
A lab-on-a-chip is a device that integrates one or several laboratory functions on a single chip of only millimeters to a few square centimeters in size. LOCs deal with the handling of extremely small fluid volumes down to less than pico liters. Lab-on-a-chip devices are a subset of MEMS devices...
application for DNA sequencing, in which the Sanger sequencing steps (thermal cycling, sample purification, and capillary electrophoresis) are integrated on a wafer-scale chip using nanoliter-scale sample volumes. This technology generates long and accurate sequence reads, while obviating many of the significant shortcomings of the conventional Sanger method (e.g. high consumption of expensive reagents, reliance on expensive equipment, personnel-intensive manipulations, etc.) by integrating and automating the Sanger sequencing steps.
Applications of Microfluidic Sequencing Technologies
Other useful applications of DNA sequencing include single nucleotide polymorphismSingle nucleotide polymorphism
A single-nucleotide polymorphism is a DNA sequence variation occurring when a single nucleotide — A, T, C or G — in the genome differs between members of a biological species or paired chromosomes in an individual...
(SNP) detection, single-strand conformation polymorphism
Single strand conformation polymorphism
Single-strand conformation polymorphism , or single-strand chain polymorphism, is defined as conformational difference of single-stranded nucleotide sequences of identical length as induced by differences in the sequences under certain experimental conditions...
(SSCP) hetroduplex analysis, and short tandem repeat
Short tandem repeat
A short tandem repeat in DNA occurs when a pattern of two or more nucleotides are repeated and the repeated sequences are directly adjacent to each other. The pattern can range in length from 2 to 5 base pairs and is typically in the non-coding intron region...
(STR) analysis. Resolving DNA fragments according to differences in size and/or conformation is the most critical step in studying these features of the genome.
Device design
A microfluidic sequencing chip developed by Richard Mathies and colleagues (University of California, Berkeley).The sequencing chip has a four-layer construction, consisting of three 100-mm-diameter glass wafers (on which device elements are microfabricated) and a polydimethylsiloxane (PDMS) membrane. Reaction chambers and capillary electrophoresis channels are etched between the top two glass wafers, which are thermally bonded. Three-dimensional channel interconnections and microvalves are formed by the PDMS and bottom manifold glass wafer.
The device consists of three functional units, each corresponding to the Sanger sequencing steps. The Thermal Cycling (TC) unit is a 250-nanoliter reaction chamber with integrated resistive temperature detector, microvalves, and a surface heater. Movement of reagent between the top all-glass layer and the lower glass-PDMS layer occurs through 500-μm-diameter via-holes. After thermal-cycling, the reaction mixture undergoes purification in the capture/purification chamber, and then is injected into the capillary electrophoresis (CE) chamber. The CE unit consists of a 30-cm capillary which is folded into a compact switchback pattern via 65-μm-wide turns.
Sequencing chemistry
- Thermal cycling
In the TC reaction chamber, dye-terminator sequencing reagent, template DNA, and primers are loaded into the TC chamber and thermal-cycled for 35 cycles ( at 95°C for 12 seconds and at 60°C for 55 seconds).
- Purification
The charged reaction mixture (containing extension fragments, template DNA, and excess sequencing reagent) is conducted through a capture/purification chamber at 30°C via a 33-Volts/cm electric field applied between capture outlet and inlet ports. The capture gel through which the sample is driven, consists of 40 μM of oligonucleotide (complementary to the primers) covalently bound to a polyacrylamide matrix. Extension fragments are immobilized by the gel matrix, and excess primer, template, free nucleotides, and salts are eluted through the capture waste port. The capture gel is heated to 67-75°C to release extension fragments.
- Capillary electrophoresis
Extension fragments are injected into the CE chamber where they are electrophoresed through a 125-167-V/cm field.
Platforms
The Apollo 100 platform (Microchip Biotechnologies Inc., Dublin, CA) integrates the first two Sanger sequencing steps (thermal cycling and purification) in a fully automated system. The manufacturer claims that samples are ready for capillary electrophoresis within three hours of the sample and reagents being loaded into the system. The Apollo 100 platform requires sub-microliter volumes of reagents.Comparisons to other sequencing techniques

Help:http://en.wikipedia.org/wiki/DNA_sequencing#Chain-termination_methods
External links
Popular Media Articles:http://nano.cancer.gov/news_center/nanotech_news_2006-05-30a.asp
http://nano.cancer.gov/news_center/monthly_feature_2005_aug.asp
http://www.genomeweb.com/sequencing/mbi-says-new-tool-automates-sanger-sample-prep-cuts-reagent-and-labor-costs
Commercial Websites:
Illumina/Solexa Website
Roche 454 Life Sciences Website
ABI SOLiD website

