Exon junction complex
Encyclopedia
The exon
junction complex (EJC) has major influences on translation, surveillance
and localization of the spliced
mRNA. It is first deposited onto mRNA during splicing and is then transported into the cytoplasm
. There it plays a major role in post-transcriptional regulation of mRNA. It is believed that exon junction complexes provide a position-specific memory of the splicing event. The EJC consists of a stable heterotetramer core, which serves as a binding platform for other factors necessary for the mRNA pathway. The core of the EJC contains the protein
eukaryotic
initiation factor
4AIII (eIF4AIII; a DEAD-box
RNA
helicase
) bound to an adenosine triphosphate (ATP
) analog, as well as the additional proteins Magoh
and Y14. In order for the binding of the complex to the mRNA to occur, the eIF4AIII factor is inhibited, stopping the hydrolysis of ATP. This recognizes EJC as an ATP dependent complex.
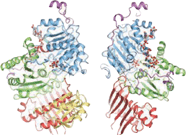
, Y14, SRm160, Aly/REF and Magoh, among others. RNPS1 can function as a coactivator of splicing, but along with Y14, it also takes part in the process of nonsense-mediated decay
in eukaryotes. SRm160 is another coactivator that has been proposed to enhance mRNA 3’ end processing. The protein component Magoh is thought to facilitate the subcytoplasmic localization of mRNAs while Aly is engaged in nuclear mRNA export. Aly is believed to be recruited to the exon junction complex by the protein UAP56. UAP56 is recognized as a RNA helicase but acts as a splicing factor required for early splicesome assembly. Another factor involved in the EJC pathway is DEK. This component is known to take part in a variety of functions ranging from splicing to transcriptional regulation and chromatin
structure.
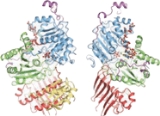
of the exon junction complex has revealed the structural organization of its protein components. The core of the complex is elongated with an overall dimension of 99Å by 67Å by 54Å. It is organized around the eIF4AIII factor. The factor itself consists of two different types of conformations around the mRNA: closed and open. In a closed state, the two domains of eIF4AIII form composite binding sites for the 5'-adenylyl-ß- -imidodiphosphate (ADPNP) and mRNA. In the open conformation, the two domains are rotated by 160 degrees relative to closed state18. The protein components Magoh and Y14 bind together to form a heterodimer located at the 5’ pole of the EJC. Magoh binds to an eIF4AIII domain through interactions between residues from its two C-terminal helices and one end of a large β-sheet
. Conserved residues in the linker between the two eIF4AIII domains form salt bridges or hydrogen bonds with specific residues in Magoh. Other bonding occurs between the second loop of the Magoh β–sheet and the two eIF4AIII domains and their linker. There is only a single partial bond formed between Y14 and eIF4AIII. This consists of a salt bridge between the conserved residues Y14 Arg108
and eIF4AIII Asp401
. If mutations were to occur to both of these residues, association of Magoh-Y14 with EJC would be non-existent.
(mRNP). The EJC remains stably bound to this mRNP as it is exported out of the nucleus and into the cytoplasm. Protein components are either bound to or released by the EJC as it is transported. In order for the translocation of mRNAs through the nuclear pore complex to occur, a heterodimer consisting of NXF1
/TAP and NXT1
/p15
must bind to the transcripts. NXF1/TAP is a major receptor for the export of mRNAs to the cytoplasm. This is because it interacts with both RNA-binding adapter proteins and components of the nuclear pore
complex.
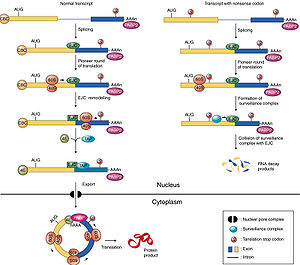
. More specifically, they are found in the nonsense mediated decay
pathway (NMD). In normal mRNA translation, the ribosome
binds to the transcript and begins amino acid
chain elongation. It continues on until it reaches the location of the exon junction complex, which it then displaces. Next, translation is complete when the ribosome reaches a termination codon. In NMD, the mRNA transcript contains a premature termination codon (PTC) due to a nonsense mutation
. If this codon occurs prior to the EJC site, then mRNA decay is triggered. The EJC and its position serve as a type of regulator, determining whether the transcript is defective or not.
EJCs are also known to take part in NMD in another way; the recruitment of the surveillance factors UPF1
, UPF2
and UPF3
. These proteins are the most important components of the NMD mechanism. The EJC protein MAGOH, Y14 and eIF4AIII provide a binding for UPF3, which acts as a bridge between UPF2 and UPF1 forming a trimeric complex. Within this complex, UPF2 and UPF3 act cooperatively to promote ATPase
and RNA helicase of UPF1. The EJC core stably anchors the UPF complex to the mRNA, and aids in regulation of essential UPF1 protein. Ribosomes which are stalled on a PTC recruit UPF1 through interactions with the release factor eRF1 and eRF3. Along with the protein SMG1
, eRF1, eRF3 and UPF1 form the complex SURF. This complex forms a bridge between the ribosome and the downstream EJC which is associated with UPF3 and UPF2. This interaction triggers the phosphorylation
of UPF1 by SMG1, causing the dissocation of eRF1 and eRF3. The complex produced consists of EJC, UPF3, UPF2, phosphorylated UPF1 and SMG1 and in turn triggers degradation of the mRNA.
Exon
An exon is a nucleic acid sequence that is represented in the mature form of an RNA molecule either after portions of a precursor RNA have been removed by cis-splicing or when two or more precursor RNA molecules have been ligated by trans-splicing. The mature RNA molecule can be a messenger RNA...
junction complex (EJC) has major influences on translation, surveillance
MRNA surveillance
mRNA surveillance mechanisms are pathways utilized by organisms to ensure fidelity and quality of messenger RNA molecules. There are a number of surveillance mechanisms present within cells...
and localization of the spliced
RNA splicing
In molecular biology and genetics, splicing is a modification of an RNA after transcription, in which introns are removed and exons are joined. This is needed for the typical eukaryotic messenger RNA before it can be used to produce a correct protein through translation...
mRNA. It is first deposited onto mRNA during splicing and is then transported into the cytoplasm
Cytoplasm
The cytoplasm is a small gel-like substance residing between the cell membrane holding all the cell's internal sub-structures , except for the nucleus. All the contents of the cells of prokaryote organisms are contained within the cytoplasm...
. There it plays a major role in post-transcriptional regulation of mRNA. It is believed that exon junction complexes provide a position-specific memory of the splicing event. The EJC consists of a stable heterotetramer core, which serves as a binding platform for other factors necessary for the mRNA pathway. The core of the EJC contains the protein
Protein
Proteins are biochemical compounds consisting of one or more polypeptides typically folded into a globular or fibrous form, facilitating a biological function. A polypeptide is a single linear polymer chain of amino acids bonded together by peptide bonds between the carboxyl and amino groups of...
eukaryotic
Eukaryote
A eukaryote is an organism whose cells contain complex structures enclosed within membranes. Eukaryotes may more formally be referred to as the taxon Eukarya or Eukaryota. The defining membrane-bound structure that sets eukaryotic cells apart from prokaryotic cells is the nucleus, or nuclear...
initiation factor
Initiation factor
Initiation factors are proteins that bind to the small subunit of the ribosome during the initiation of translation, a part of protein biosynthesis.They are divided into three major groups:*Prokaryotic initiation factors*Archaeal initiation factors...
4AIII (eIF4AIII; a DEAD-box
DEAD box
DEAD box proteins are involved in an assortment of metabolic processes that involve RNA. They are highly conserved in nine domains and can be found in both prokaryotes and eukaryotes, but not all...
RNA
RNA
Ribonucleic acid , or RNA, is one of the three major macromolecules that are essential for all known forms of life....
helicase
Helicase
Helicases are a class of enzymes vital to all living organisms. They are motor proteins that move directionally along a nucleic acid phosphodiester backbone, separating two annealed nucleic acid strands using energy derived from ATP hydrolysis.-Function:Many cellular processes Helicases are a...
) bound to an adenosine triphosphate (ATP
Adenosine triphosphate
Adenosine-5'-triphosphate is a multifunctional nucleoside triphosphate used in cells as a coenzyme. It is often called the "molecular unit of currency" of intracellular energy transfer. ATP transports chemical energy within cells for metabolism...
) analog, as well as the additional proteins Magoh
MAGOH
Protein mago nashi homolog is a protein that in humans is encoded by the MAGOH gene.-Interactions:MAGOH has been shown to interact with RBM8A and NXF1.-Further reading:...
and Y14. In order for the binding of the complex to the mRNA to occur, the eIF4AIII factor is inhibited, stopping the hydrolysis of ATP. This recognizes EJC as an ATP dependent complex.

Protein components
The EJC is made up of several key protein components: RNPS1RNPS1
RNA-binding protein with serine-rich domain 1 is a protein that in humans is encoded by the RNPS1 gene.-Interactions:RNPS1 has been shown to interact with SART3 and Pinin.-Further reading:...
, Y14, SRm160, Aly/REF and Magoh, among others. RNPS1 can function as a coactivator of splicing, but along with Y14, it also takes part in the process of nonsense-mediated decay
Nonsense mediated decay
Nonsense-mediated decay is a cellular mechanism of mRNA surveillance that functions to detect nonsense mutations and prevent the expression of truncated or erroneous proteins. Following transcription, precursor mRNA undergoes an assemblage of ribonucleoprotein components followed by regulatory...
in eukaryotes. SRm160 is another coactivator that has been proposed to enhance mRNA 3’ end processing. The protein component Magoh is thought to facilitate the subcytoplasmic localization of mRNAs while Aly is engaged in nuclear mRNA export. Aly is believed to be recruited to the exon junction complex by the protein UAP56. UAP56 is recognized as a RNA helicase but acts as a splicing factor required for early splicesome assembly. Another factor involved in the EJC pathway is DEK. This component is known to take part in a variety of functions ranging from splicing to transcriptional regulation and chromatin
Chromatin
Chromatin is the combination of DNA and proteins that make up the contents of the nucleus of a cell. The primary functions of chromatin are; to package DNA into a smaller volume to fit in the cell, to strengthen the DNA to allow mitosis and meiosis and prevent DNA damage, and to control gene...
structure.
Structure
The crystallizationCrystallization
Crystallization is the process of formation of solid crystals precipitating from a solution, melt or more rarely deposited directly from a gas. Crystallization is also a chemical solid–liquid separation technique, in which mass transfer of a solute from the liquid solution to a pure solid...
of the exon junction complex has revealed the structural organization of its protein components. The core of the complex is elongated with an overall dimension of 99Å by 67Å by 54Å. It is organized around the eIF4AIII factor. The factor itself consists of two different types of conformations around the mRNA: closed and open. In a closed state, the two domains of eIF4AIII form composite binding sites for the 5'-adenylyl-ß- -imidodiphosphate (ADPNP) and mRNA. In the open conformation, the two domains are rotated by 160 degrees relative to closed state18. The protein components Magoh and Y14 bind together to form a heterodimer located at the 5’ pole of the EJC. Magoh binds to an eIF4AIII domain through interactions between residues from its two C-terminal helices and one end of a large β-sheet
Beta sheet
The β sheet is the second form of regular secondary structure in proteins, only somewhat less common than the alpha helix. Beta sheets consist of beta strands connected laterally by at least two or three backbone hydrogen bonds, forming a generally twisted, pleated sheet...
. Conserved residues in the linker between the two eIF4AIII domains form salt bridges or hydrogen bonds with specific residues in Magoh. Other bonding occurs between the second loop of the Magoh β–sheet and the two eIF4AIII domains and their linker. There is only a single partial bond formed between Y14 and eIF4AIII. This consists of a salt bridge between the conserved residues Y14 Arg108
Arginine
Arginine is an α-amino acid. The L-form is one of the 20 most common natural amino acids. At the level of molecular genetics, in the structure of the messenger ribonucleic acid mRNA, CGU, CGC, CGA, CGG, AGA, and AGG, are the triplets of nucleotide bases or codons that codify for arginine during...
and eIF4AIII Asp401
Asparagine
Asparagine is one of the 20 most common natural amino acids on Earth. It has carboxamide as the side-chain's functional group. It is not an essential amino acid...
. If mutations were to occur to both of these residues, association of Magoh-Y14 with EJC would be non-existent.
Mechanism
During the second step of splicing in eukaryotic cells, the EJC is deposited approximately 20-24 nucleotides from the 5’ end upstream of the splice junction (where two exons are joined), when the lariat has formed and the exons are ligated together. The binding of the EJC to the mRNA occurs in a sequence independent manner, to form the mature messenger ribonucleoproteinRibonucleoprotein
Ribonucleoprotein is a nucleoprotein that contains RNA, i.e. it is an association that combines ribonucleic acid and protein together. A few known examples include the ribosome, the enzyme telomerase, vault ribonucleoproteins, and small nuclear RNPs , which are implicated in pre-mRNA splicing and...
(mRNP). The EJC remains stably bound to this mRNP as it is exported out of the nucleus and into the cytoplasm. Protein components are either bound to or released by the EJC as it is transported. In order for the translocation of mRNAs through the nuclear pore complex to occur, a heterodimer consisting of NXF1
NXF1
Nuclear RNA export factor 1, also known as NXF1, is a protein which in humans is encoded by the NXF1 gene.-Function:This gene is one member of a family of nuclear RNA export factor genes...
/TAP and NXT1
NXT1
NTF2-related export protein 1 is a protein that in humans is encoded by the NXT1 gene.-Further reading:...
/p15
CDKN2B
Cyclin-dependent kinase 4 inhibitor B also known as multiple tumor suppressor 2 or p15INK4B is a protein that is encoded by the CDKN2B gene in humans.- Function :...
must bind to the transcripts. NXF1/TAP is a major receptor for the export of mRNAs to the cytoplasm. This is because it interacts with both RNA-binding adapter proteins and components of the nuclear pore
Nuclear pore
Nuclear pores are large protein complexes that cross the nuclear envelope, which is the double membrane surrounding the eukaryotic cell nucleus. There are about on average 2000 nuclear pore complexes in the nuclear envelope of a vertebrate cell, but it varies depending on cell type and the stage in...
complex.

EJC in nonsense mediated decay
Exon junction complexes play a major role in mRNA surveillanceMRNA surveillance
mRNA surveillance mechanisms are pathways utilized by organisms to ensure fidelity and quality of messenger RNA molecules. There are a number of surveillance mechanisms present within cells...
. More specifically, they are found in the nonsense mediated decay
Nonsense mediated decay
Nonsense-mediated decay is a cellular mechanism of mRNA surveillance that functions to detect nonsense mutations and prevent the expression of truncated or erroneous proteins. Following transcription, precursor mRNA undergoes an assemblage of ribonucleoprotein components followed by regulatory...
pathway (NMD). In normal mRNA translation, the ribosome
Ribosome
A ribosome is a component of cells that assembles the twenty specific amino acid molecules to form the particular protein molecule determined by the nucleotide sequence of an RNA molecule....
binds to the transcript and begins amino acid
Amino acid
Amino acids are molecules containing an amine group, a carboxylic acid group and a side-chain that varies between different amino acids. The key elements of an amino acid are carbon, hydrogen, oxygen, and nitrogen...
chain elongation. It continues on until it reaches the location of the exon junction complex, which it then displaces. Next, translation is complete when the ribosome reaches a termination codon. In NMD, the mRNA transcript contains a premature termination codon (PTC) due to a nonsense mutation
Nonsense mutation
In genetics, a nonsense mutation is a point mutation in a sequence of DNA that results in a premature stop codon, or a nonsense codon in the transcribed mRNA, and in a truncated, incomplete, and usually nonfunctional protein product. It differs from a missense mutation, which is a point mutation...
. If this codon occurs prior to the EJC site, then mRNA decay is triggered. The EJC and its position serve as a type of regulator, determining whether the transcript is defective or not.
EJCs are also known to take part in NMD in another way; the recruitment of the surveillance factors UPF1
UPF1
Regulator of nonsense transcripts 1 is a protein that in humans is encoded by the UPF1 gene.-Interactions:UPF1 has been shown to interact with UPF2, SMG1, DCP2, DCP1A, UPF3A and UPF3B.-Further reading:...
, UPF2
UPF2
Regulator of nonsense transcripts 2 is a protein that in humans is encoded by the UPF2 gene.-Interactions:UPF2 has been shown to interact with UPF1, UPF3A and UPF3B.-Further reading:...
and UPF3
UPF3B
Regulator of nonsense transcripts 3B is a protein that in humans is encoded by the UPF3B gene.-Interactions:UPF3B has been shown to interact with UPF2 and UPF1.-Further reading:...
. These proteins are the most important components of the NMD mechanism. The EJC protein MAGOH, Y14 and eIF4AIII provide a binding for UPF3, which acts as a bridge between UPF2 and UPF1 forming a trimeric complex. Within this complex, UPF2 and UPF3 act cooperatively to promote ATPase
ATPase
ATPases are a class of enzymes that catalyze the decomposition of adenosine triphosphate into adenosine diphosphate and a free phosphate ion. This dephosphorylation reaction releases energy, which the enzyme harnesses to drive other chemical reactions that would not otherwise occur...
and RNA helicase of UPF1. The EJC core stably anchors the UPF complex to the mRNA, and aids in regulation of essential UPF1 protein. Ribosomes which are stalled on a PTC recruit UPF1 through interactions with the release factor eRF1 and eRF3. Along with the protein SMG1
SMG1 (gene)
Serine/threonine-protein kinase SMG1 is an enzyme that in humans is encoded by the SMG1 gene. SMG1 belongs to the phosphatidylinositol 3-kinase-related kinase protein family.-Interactions:SMG1 has been shown to interact with PRKCI and UPF1....
, eRF1, eRF3 and UPF1 form the complex SURF. This complex forms a bridge between the ribosome and the downstream EJC which is associated with UPF3 and UPF2. This interaction triggers the phosphorylation
Phosphorylation
Phosphorylation is the addition of a phosphate group to a protein or other organic molecule. Phosphorylation activates or deactivates many protein enzymes....
of UPF1 by SMG1, causing the dissocation of eRF1 and eRF3. The complex produced consists of EJC, UPF3, UPF2, phosphorylated UPF1 and SMG1 and in turn triggers degradation of the mRNA.

