
E2F
Encyclopedia
E2F is a group of genes that codifies a family of transcription factor
s (TF) in higher eukaryotes. Three of them are activators: E2F1, 2 and E2F3a. Six others act as suppressors: E2F3b, E2F4-8. All of them are involved in the cell cycle
regulation and synthesis of DNA in mammal
ian cells. E2Fs as TFs bind to the TTTCGCGC consensus binding site
in the target promoter sequence.
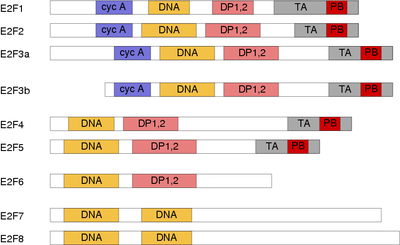
s of E2F family members (N-terminus to the right, C-terminus to the left) highlighting the relative locations of functional domains
within each member:
E2F1 protein sequences from NCBI
protein and nucleotide database.
analysis has shown that the E2F family of transcription factors has a fold similar to the winged-helix
DNA-binding motif
.
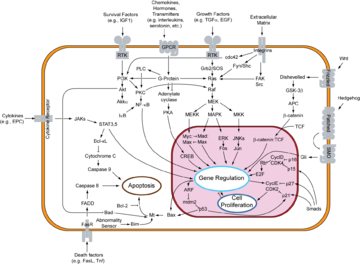 E2F family members play a major role during the G1/S transition in mammalian and plant cell cycle
E2F family members play a major role during the G1/S transition in mammalian and plant cell cycle
(see KEGG cell cycle pathway). DNA microarray
analysis reveals unique sets of target promoters among E2F family members suggesting that each protein has a unique role in the cell cycle. Among E2F transcriptional targets are cyclin
s, cdk
s, checkpoints regulators, DNA repair
and replication proteins. Nonetheless, there is a great deal of redundancy among the family members. Mouse embryos lacking E2F1, E2F2, and one of the E2F3 isoforms, can develop normally when either E2F3a or E2F3b, is expressed.
The E2F family is generally split by function into two groups: transcription activators and repressors. Activators such as E2F1, E2F2, E2F3a promote and help carryout the cell cycle, while repressors inhibit the cell cycle. Yet, both sets of E2F have similar domains. E2F1-6 have DP1,2 heterodimerization domain which allows them to bind to DP1 or DP2, proteins distantly related to E2F. Binding with DP1,2 provides a second DNA binding site, increasing E2F binding stability. Most E2F have a pocket protein binding domain. Pocket proteins such as pRB
and related proteins p107 and p130, can bind to E2F when hypophosphorylated. In activators, E2F binding with pRB has been shown to mask the transactivation domain responsible for transcription activation. In repressors E2F4 and E2F5, pocket protein binding (more often p107 and p130 than pRB) mediates recruitment of repression complexes to silence target genes. E2F6, E2F7, and E2F8 do not have pocket protein binding sites and their mechanism for gene silencing is unclear. Cdk4(6)/cyclin D and cdk2/cyclin E phosphorylate pRB and related pocket proteins allowing them to disassociate from E2F. Activator E2F proteins can then transcribe S phase promoting genes. In REF52 cells, overexpression of activator E2F1 is able to push quiescent cells into S phase. While repressors E2F4 and 5 do not alter cell proliferation, they mediate G1 arrest.
E2F activator levels are cyclic, with maximal expression during G1/S. In contrast, E2F repressors stay constant, especially since they are often expressed in quiescent cells. Specifically, E2F5 is only expressed in terminally differentiated cells in mice. The balance between repressor and activator E2F regulate cell cycle progression. When activator E2F family proteins are knocked out, repressors become active to inhibit E2F target genes.
(pRb) binds to the E2F-1 transcription factor preventing it from interacting with the cell's transcription machinery. In the absence of pRb, E2F-1 (along with its binding partner DP-1) mediates the trans-activation of E2F-1 target genes that facilitate the G1/S transition and S-phase. E2F target genes encode proteins involved in DNA replication (for example DNA polymerase, thymidine kinase (TK), dihydrofolate reductase (DHFR) and cdc6), chromosomal replication (replication origin-binding protein HsOrc1 and MCM 5). When cells are not proliferating, E2F DNA binding sites contribute to transcriptional repression. In vivo footprinting experiments obtained on Cdc2 and B-myb promoters demonstrated E2F DNA binding site occupation during G0 and early G1, when E2F is in transcriptional repressive complexes with the pocket proteins.
pRb is one of the targets of the oncogenic Human Papilloma Virus protein E7. By binding to pRB it stops the regulation of E2F transcription factors and cancer can proceed.
stimulation and the subsequent phosphorylation
of the E2F inhibitor Retinoblastoma protein, pRB
. The phosphorylation
of pRB is initiated by Cyclin D
/cdk4, CDK6 complex and continued by Cyclin E/cdk2. Cyclin D/cdk4,6 itself is activated by the MAPK signaling pathway.
When bound to E2F-3a, pRb can directly repress E2F-3a target genes by recruiting chromatin remodeling complexes and histone modifying activities (e.g. histone deacetylase, HDAC) to the promoter.
Transcription factor
In molecular biology and genetics, a transcription factor is a protein that binds to specific DNA sequences, thereby controlling the flow of genetic information from DNA to mRNA...
s (TF) in higher eukaryotes. Three of them are activators: E2F1, 2 and E2F3a. Six others act as suppressors: E2F3b, E2F4-8. All of them are involved in the cell cycle
Cell cycle
The cell cycle, or cell-division cycle, is the series of events that takes place in a cell leading to its division and duplication . In cells without a nucleus , the cell cycle occurs via a process termed binary fission...
regulation and synthesis of DNA in mammal
Mammal
Mammals are members of a class of air-breathing vertebrate animals characterised by the possession of endothermy, hair, three middle ear bones, and mammary glands functional in mothers with young...
ian cells. E2Fs as TFs bind to the TTTCGCGC consensus binding site
Binding site
In biochemistry, a binding site is a region on a protein, DNA, or RNA to which specific other molecules and ions—in this context collectively called ligands—form a chemical bond...
in the target promoter sequence.
E2F family
Schematic diagram of the amino acid sequencePeptide sequence
Peptide sequence or amino acid sequence is the order in which amino acid residues, connected by peptide bonds, lie in the chain in peptides and proteins. The sequence is generally reported from the N-terminal end containing free amino group to the C-terminal end containing free carboxyl group...
s of E2F family members (N-terminus to the right, C-terminus to the left) highlighting the relative locations of functional domains
Protein domain
A protein domain is a part of protein sequence and structure that can evolve, function, and exist independently of the rest of the protein chain. Each domain forms a compact three-dimensional structure and often can be independently stable and folded. Many proteins consist of several structural...
within each member:
| Family members | Legend |
|---|---|
 |
|
Genes
Homo sapiens E2F1 mRNA orE2F1 protein sequences from NCBI
National Center for Biotechnology Information
The National Center for Biotechnology Information is part of the United States National Library of Medicine , a branch of the National Institutes of Health. The NCBI is located in Bethesda, Maryland and was founded in 1988 through legislation sponsored by Senator Claude Pepper...
protein and nucleotide database.
Structure
X-ray crystallographicX-ray crystallography
X-ray crystallography is a method of determining the arrangement of atoms within a crystal, in which a beam of X-rays strikes a crystal and causes the beam of light to spread into many specific directions. From the angles and intensities of these diffracted beams, a crystallographer can produce a...
analysis has shown that the E2F family of transcription factors has a fold similar to the winged-helix
Winged-helix transcription factors
Consisting of about 110 amino acids, the domain in winged-helix transcription factors has four helices and a two-strand beta-sheet.These proteins are classified into 17 families called FoxA-FoxQ....
DNA-binding motif
DNA-binding domain
A DNA-binding domain is an independently folded protein domain that contains at least one motif that recognizes double- or single-stranded DNA. A DBD can recognize a specific DNA sequence or have a general affinity to DNA...
.
Role in the cell cycle

Cell cycle
The cell cycle, or cell-division cycle, is the series of events that takes place in a cell leading to its division and duplication . In cells without a nucleus , the cell cycle occurs via a process termed binary fission...
(see KEGG cell cycle pathway). DNA microarray
DNA microarray
A DNA microarray is a collection of microscopic DNA spots attached to a solid surface. Scientists use DNA microarrays to measure the expression levels of large numbers of genes simultaneously or to genotype multiple regions of a genome...
analysis reveals unique sets of target promoters among E2F family members suggesting that each protein has a unique role in the cell cycle. Among E2F transcriptional targets are cyclin
Cyclin
Cyclins are a family of proteins that control the progression of cells through the cell cycle by activating cyclin-dependent kinase enzymes.- Function :...
s, cdk
Cyclin-dependent kinase
thumb|350px|Schematic of the cell cycle. outer ring: I=[[Interphase]], M=[[Mitosis]]; inner ring: M=Mitosis; G1=[[G1 phase|Gap phase 1]]; S=[[S phase|Synthesis]]; G2=[[G2 phase|Gap phase 2]]...
s, checkpoints regulators, DNA repair
DNA repair
DNA repair refers to a collection of processes by which a cell identifies and corrects damage to the DNA molecules that encode its genome. In human cells, both normal metabolic activities and environmental factors such as UV light and radiation can cause DNA damage, resulting in as many as 1...
and replication proteins. Nonetheless, there is a great deal of redundancy among the family members. Mouse embryos lacking E2F1, E2F2, and one of the E2F3 isoforms, can develop normally when either E2F3a or E2F3b, is expressed.
The E2F family is generally split by function into two groups: transcription activators and repressors. Activators such as E2F1, E2F2, E2F3a promote and help carryout the cell cycle, while repressors inhibit the cell cycle. Yet, both sets of E2F have similar domains. E2F1-6 have DP1,2 heterodimerization domain which allows them to bind to DP1 or DP2, proteins distantly related to E2F. Binding with DP1,2 provides a second DNA binding site, increasing E2F binding stability. Most E2F have a pocket protein binding domain. Pocket proteins such as pRB
Retinoblastoma protein
The retinoblastoma protein is a tumor suppressor protein that is dysfunctional in the majority types of cancer. One highly studied function of pRb is to prevent excessive cell growth by inhibiting cell cycle progression until a cell is ready to divide...
and related proteins p107 and p130, can bind to E2F when hypophosphorylated. In activators, E2F binding with pRB has been shown to mask the transactivation domain responsible for transcription activation. In repressors E2F4 and E2F5, pocket protein binding (more often p107 and p130 than pRB) mediates recruitment of repression complexes to silence target genes. E2F6, E2F7, and E2F8 do not have pocket protein binding sites and their mechanism for gene silencing is unclear. Cdk4(6)/cyclin D and cdk2/cyclin E phosphorylate pRB and related pocket proteins allowing them to disassociate from E2F. Activator E2F proteins can then transcribe S phase promoting genes. In REF52 cells, overexpression of activator E2F1 is able to push quiescent cells into S phase. While repressors E2F4 and 5 do not alter cell proliferation, they mediate G1 arrest.
E2F activator levels are cyclic, with maximal expression during G1/S. In contrast, E2F repressors stay constant, especially since they are often expressed in quiescent cells. Specifically, E2F5 is only expressed in terminally differentiated cells in mice. The balance between repressor and activator E2F regulate cell cycle progression. When activator E2F family proteins are knocked out, repressors become active to inhibit E2F target genes.
E2F/pRb complexes
The Rb tumour suppressor proteinRetinoblastoma protein
The retinoblastoma protein is a tumor suppressor protein that is dysfunctional in the majority types of cancer. One highly studied function of pRb is to prevent excessive cell growth by inhibiting cell cycle progression until a cell is ready to divide...
(pRb) binds to the E2F-1 transcription factor preventing it from interacting with the cell's transcription machinery. In the absence of pRb, E2F-1 (along with its binding partner DP-1) mediates the trans-activation of E2F-1 target genes that facilitate the G1/S transition and S-phase. E2F target genes encode proteins involved in DNA replication (for example DNA polymerase, thymidine kinase (TK), dihydrofolate reductase (DHFR) and cdc6), chromosomal replication (replication origin-binding protein HsOrc1 and MCM 5). When cells are not proliferating, E2F DNA binding sites contribute to transcriptional repression. In vivo footprinting experiments obtained on Cdc2 and B-myb promoters demonstrated E2F DNA binding site occupation during G0 and early G1, when E2F is in transcriptional repressive complexes with the pocket proteins.
pRb is one of the targets of the oncogenic Human Papilloma Virus protein E7. By binding to pRB it stops the regulation of E2F transcription factors and cancer can proceed.
Activators: E2F1, E2F2, E2F3a
Activators are maximally expressed late in G1 and can be found in association with E2F regulated promoters during the G1/S transition. The activation of E2F-3a genes follows upon the growth factorGrowth factor
A growth factor is a naturally occurring substance capable of stimulating cellular growth, proliferation and cellular differentiation. Usually it is a protein or a steroid hormone. Growth factors are important for regulating a variety of cellular processes....
stimulation and the subsequent phosphorylation
Phosphorylation
Phosphorylation is the addition of a phosphate group to a protein or other organic molecule. Phosphorylation activates or deactivates many protein enzymes....
of the E2F inhibitor Retinoblastoma protein, pRB
Retinoblastoma protein
The retinoblastoma protein is a tumor suppressor protein that is dysfunctional in the majority types of cancer. One highly studied function of pRb is to prevent excessive cell growth by inhibiting cell cycle progression until a cell is ready to divide...
. The phosphorylation
Phosphorylation
Phosphorylation is the addition of a phosphate group to a protein or other organic molecule. Phosphorylation activates or deactivates many protein enzymes....
of pRB is initiated by Cyclin D
Cyclin D
Cyclin D is a member of the cyclin protein family that is involved in regulating cell cycle progression. The synthesis of cyclin D is initiated during G1 and drives the G1/S phase transition...
/cdk4, CDK6 complex and continued by Cyclin E/cdk2. Cyclin D/cdk4,6 itself is activated by the MAPK signaling pathway.
When bound to E2F-3a, pRb can directly repress E2F-3a target genes by recruiting chromatin remodeling complexes and histone modifying activities (e.g. histone deacetylase, HDAC) to the promoter.
Inhibitors: E2F3b, E2F4, E2F5, E2F6, E2F7, E2F8
- E2F3b, E2F4, E2F5 are expressed in quiescent cells and can be found associated with E2F-binding elements on E2F-target promoters during G0-phase. E2F-4 and 5 preferentially bind to p107/p130.
- E2F-6 acts as a transcriptional repressor, but through a distinct, pocket protein independent manner. E2F-6 mediates repression by direct binding to polycomb-group proteinsPolycomb-group proteinsPolycomb-group proteins are a family of proteins first discovered in fruit flies that can remodel chromatin such that epigenetic silencing of genes takes place...
or via the formation of a large multimeric complex containing Mga and Max proteins.
- E2F7 and E2F8 proteins can function as repressors independently of DP interaction. They are unique in having a duplicated conserved E2F-like DNA-binding domain and in lacking a DP1,2-dimerization domain. The exact mechanism of mediating repression is not yet understood.
Transcriptional targets
- Cell cycle: CCNA1,2, CCND1,2, CDK2Cyclin-dependent kinasethumb|350px|Schematic of the cell cycle. outer ring: I=[[Interphase]], M=[[Mitosis]]; inner ring: M=Mitosis; G1=[[G1 phase|Gap phase 1]]; S=[[S phase|Synthesis]]; G2=[[G2 phase|Gap phase 2]]...
, MYB, E2F1,2,3, TFDP1, CDC25A - Negative regulators: E2F7, RB1Retinoblastoma proteinThe retinoblastoma protein is a tumor suppressor protein that is dysfunctional in the majority types of cancer. One highly studied function of pRb is to prevent excessive cell growth by inhibiting cell cycle progression until a cell is ready to divide...
, TP107, TP21 - Checkpoints: TP53, BRCA1BRCA1BRCA1 is a human caretaker gene that produces a protein called breast cancer type 1 susceptibility protein, responsible for repairing DNA. The first evidence for the existence of the gene was provided by the King laboratory at UC Berkeley in 1990...
,2, BUB1 - Apoptosis: TP73, APAF1, CASP3CaspaseCaspases, or cysteine-aspartic proteases or cysteine-dependent aspartate-directed proteases are a family of cysteine proteases that play essential roles in apoptosis , necrosis, and inflammation....
,7,8, MAP3K5,14 - Nucleotide synthesis: thymidine kinaseThymidine kinaseThymidine kinase is an enzyme, a phosphotransferase : 2'-deoxythymidine kinase, ATP-thymidine 5'-phosphotransferase, . It can be found in most living cells. It is present in two forms in mammalian cells, TK1 and TK2...
(tk), thymidylate synthase (ts), DHFRDihydrofolate reductase- Function :Dihydrofolate reductase converts dihydrofolate into tetrahydrofolate, a methyl group shuttle required for the de novo synthesis of purines, thymidylic acid, and certain amino acids... - DNA repair: BARD1, RAD51, UNG1,2, FANCA, FANCC, FANCJ
- DNA replication: PCNAPCNAProliferating Cell Nuclear Antigen, commonly known as PCNA, is a protein that acts as a processivity factor for DNA polymerase δ in eukaryotic cells. It achieves this processivity by encircling the DNA, thus creating a topological link to the genome...
, histoneHistoneIn biology, histones are highly alkaline proteins found in eukaryotic cell nuclei that package and order the DNA into structural units called nucleosomes. They are the chief protein components of chromatin, acting as spools around which DNA winds, and play a role in gene regulation...
H2A, DNA polDNA polymeraseA DNA polymerase is an enzyme that helps catalyze in the polymerization of deoxyribonucleotides into a DNA strand. DNA polymerases are best known for their feedback role in DNA replication, in which the polymerase "reads" an intact DNA strand as a template and uses it to synthesize the new strand.... and
and  , RPA1,2,3, CDC6, MCM2,3,4,5,6,7
, RPA1,2,3, CDC6, MCM2,3,4,5,6,7
External links
- Drosophila E2F transcription factor - The Interactive Fly
- Drosophila E2F transcription factor 2 - The Interactive Fly

