Cell-free protein array
Encyclopedia
Cell-free protein array technology produces protein microarray
s by performing in vitro
synthesis of the target proteins from their DNA
templates. This method of synthesizing protein microarrays overcomes the many obstacles and challenges faced by traditional methods of protein array production that have prevented widespread adoption of protein microarrays in proteomics
. Protein arrays made from this technology can be used for testing protein–protein interactions, as well as protein interactions with other cellular molecules such as DNA and lipids. Other applications include enzymatic inhibition assays and screenings of antibody specificity.
s has generated much enthusiasm for protein microarrays. However, protein microarrays have not quite taken off as expected, even with the necessary tools and know-how from DNA microarrays being in place and ready for adaptation. One major reason is that protein microarrays are much more laborious and technically challenging to construct than DNA microarrays.
The traditional methods of producing protein arrays require the separate in vivo
expression of hundreds or thousands of proteins, followed by separate purification and immobilization of the proteins on a solid surface. Cell-free protein array technology attempts to simplify protein microarray construction by bypassing the need to express the proteins in bacteria
cells and the subsequent need to purify them. It takes advantage of available cell-free protein synthesis
technology which has demonstrated that protein synthesis can occur without an intact cell as long as cell extracts containing the DNA template, transcription
and translation raw materials and machinery are provided. Common sources of cell extracts used in cell-free protein array technology include wheat germ, Escherichia coli
, and rabbit reticulocyte
. Cell extracts from other sources such as hyperthermophile
s, hybridoma
s, Xenopus
oocyte
s, insect, mammalian and human cells have also been used.
The target proteins are synthesized in situ
on the protein microarray, directly from the DNA template, thus skipping many of the steps in traditional protein microarray production and their accompanying technical limitations. More importantly, the expression of the proteins can be done in parallel, meaning all the proteins can be expressed together in a single reaction. This ability to multiplex protein expression is a major time-saver in the production process.
. Once the newly synthesized proteins are released from the ribosome
, the tag sequence
that is also synthesized at the N- or C-terminus of each nascent protein will be bound by the capture reagent or antibody, thus immobilizing the proteins to form an array. Commonly used tags include polyhistidine
(His)6 and glutathione s-transferase (GST).
Various research groups have developed their own methods, each differing in their approach, but can be summarized into 3 main groups.
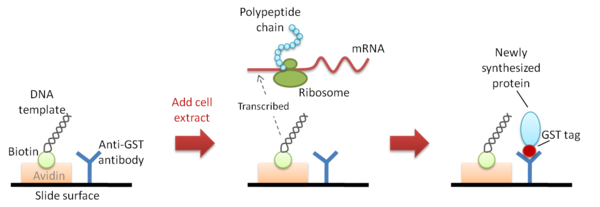 Nucleic acid programmable protein array (NAPPA): NAPPA uses DNA template that has already been immobilized onto the same protein capture surface. The DNA template is biotinylated
Nucleic acid programmable protein array (NAPPA): NAPPA uses DNA template that has already been immobilized onto the same protein capture surface. The DNA template is biotinylated
and is bound to avidin
that is pre-coated onto the protein capture surface. Newly synthesized proteins which are tagged with GST are then immobilized next to the template DNA by binding to the adjacent polyclonal anti-GST capture antibody that is also pre-coated onto the capture surface (Figure 1). The main drawback of this method is the extra and tedious preparation steps at the beginning of the process: (1) the cloning
of cDNAs in an expression-ready vector
; and (2) the need to biotinylate the plasmid
DNA but not to interfere with transcription. Moreover, the resulting protein array is not ‘pure’ because the proteins are co-localized with their DNA templates and capture antibodies.
 Protein in situ array (PISA): Unlike NAPPA, PISA completely bypasses DNA immobilization as the DNA template is added as a free molecule in the reaction mixture. In 2006, another group refined and miniaturized this method by using multiple spotting technique to spot the DNA template and cell-free transcription and translation mixture on a high-density protein microarray with up to 13,000 spots (Figure 2). This was made possible by the automated system used to accurately and sequentially supply the reagents for the transcription/translation reaction occurs in a small, sub-nanolitre droplet.
Protein in situ array (PISA): Unlike NAPPA, PISA completely bypasses DNA immobilization as the DNA template is added as a free molecule in the reaction mixture. In 2006, another group refined and miniaturized this method by using multiple spotting technique to spot the DNA template and cell-free transcription and translation mixture on a high-density protein microarray with up to 13,000 spots (Figure 2). This was made possible by the automated system used to accurately and sequentially supply the reagents for the transcription/translation reaction occurs in a small, sub-nanolitre droplet.
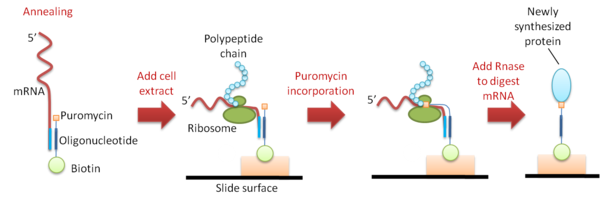 In situ puromycin-capture: This method is an adaptation of mRNA display
In situ puromycin-capture: This method is an adaptation of mRNA display
technology. PCR DNA is first transcribed to mRNA, and a single-stranded DNA oligonucleotide
modified with biotin
and puromycin
on each end is then hybridized to the 3’-end of the mRNA. The mRNAs are then arrayed on a slide and immobilized by the binding of biotin to streptavidin
that is pre-coated on the slide. Cell extract is then dispensed on the slide for in situ translation to take place. When the ribosome reaches the hybridized oligonucleotide, it stalls and incorporates the puromycin molecule to the nascent polypeptide chain, thereby attaching the newly synthesized protein to the microarray via the DNA oligonucleotide (Figure 3). A pure protein array is obtained after the mRNA is digested with RNase. The protein spots generated by this method are very sharply defined and can be produced at a high density.
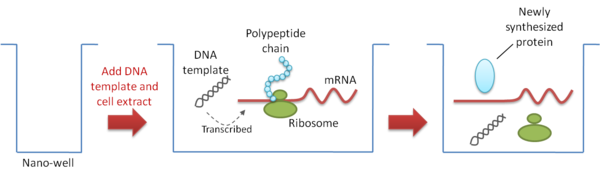 Nano-well array formats are used to express individual proteins in small volume reaction vessels or nano-wells (Figure 4). This format is sometimes preferred because it avoids the need to immobilize the target protein which might result in the potential loss of protein activity. The miniaturization of the array also conserves solution and precious compounds that might be used in screening assays. Moreover, the structural properties of individual wells help to prevent cross-contamination among chambers.
Nano-well array formats are used to express individual proteins in small volume reaction vessels or nano-wells (Figure 4). This format is sometimes preferred because it avoids the need to immobilize the target protein which might result in the potential loss of protein activity. The miniaturization of the array also conserves solution and precious compounds that might be used in screening assays. Moreover, the structural properties of individual wells help to prevent cross-contamination among chambers.
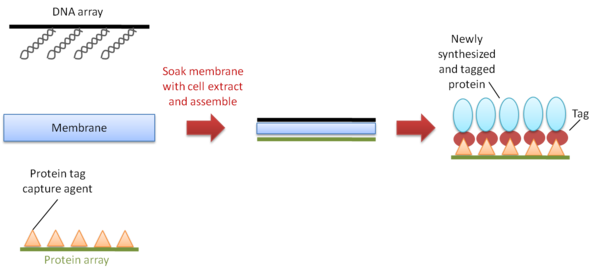 DNA array to protein array (DAPA) is a method developed in 2007 to repeatedly produce protein arrays by ‘printing’ them from a single DNA template array, on demand (Figure 5). It starts with the spotting and immobilization of an array of DNA templates onto a glass slide. The slide is then assembled face-to-face with a second slide pre-coated with a protein-capturing reagent, and a membrane soaked with cell extract is placed between the two slides for transcription and translation to take place. The newly-synthesized his-tagged proteins are then immobilized onto the slide to form the array. Over 20 protein arrays can be printed from a single DNA array with no adverse effects on production efficiency.
DNA array to protein array (DAPA) is a method developed in 2007 to repeatedly produce protein arrays by ‘printing’ them from a single DNA template array, on demand (Figure 5). It starts with the spotting and immobilization of an array of DNA templates onto a glass slide. The slide is then assembled face-to-face with a second slide pre-coated with a protein-capturing reagent, and a membrane soaked with cell extract is placed between the two slides for transcription and translation to take place. The newly-synthesized his-tagged proteins are then immobilized onto the slide to form the array. Over 20 protein arrays can be printed from a single DNA array with no adverse effects on production efficiency.
Rapid and cost-effective:
Improves protein availability:
Enables long term storage
Flexible
Protein microarray
A protein microarray, sometimes referred to as a protein binding microarray,provides a multiplex approach to identify protein–protein interactions, to identify the substrates of protein kinases, to identify transcription factor protein-activation, or to identify the targets of biologically active...
s by performing in vitro
In vitro
In vitro refers to studies in experimental biology that are conducted using components of an organism that have been isolated from their usual biological context in order to permit a more detailed or more convenient analysis than can be done with whole organisms. Colloquially, these experiments...
synthesis of the target proteins from their DNA
DNA
Deoxyribonucleic acid is a nucleic acid that contains the genetic instructions used in the development and functioning of all known living organisms . The DNA segments that carry this genetic information are called genes, but other DNA sequences have structural purposes, or are involved in...
templates. This method of synthesizing protein microarrays overcomes the many obstacles and challenges faced by traditional methods of protein array production that have prevented widespread adoption of protein microarrays in proteomics
Proteomics
Proteomics is the large-scale study of proteins, particularly their structures and functions. Proteins are vital parts of living organisms, as they are the main components of the physiological metabolic pathways of cells. The term "proteomics" was first coined in 1997 to make an analogy with...
. Protein arrays made from this technology can be used for testing protein–protein interactions, as well as protein interactions with other cellular molecules such as DNA and lipids. Other applications include enzymatic inhibition assays and screenings of antibody specificity.
Overview / background
The runaway success of DNA microarrayDNA microarray
A DNA microarray is a collection of microscopic DNA spots attached to a solid surface. Scientists use DNA microarrays to measure the expression levels of large numbers of genes simultaneously or to genotype multiple regions of a genome...
s has generated much enthusiasm for protein microarrays. However, protein microarrays have not quite taken off as expected, even with the necessary tools and know-how from DNA microarrays being in place and ready for adaptation. One major reason is that protein microarrays are much more laborious and technically challenging to construct than DNA microarrays.
The traditional methods of producing protein arrays require the separate in vivo
In vivo
In vivo is experimentation using a whole, living organism as opposed to a partial or dead organism, or an in vitro controlled environment. Animal testing and clinical trials are two forms of in vivo research...
expression of hundreds or thousands of proteins, followed by separate purification and immobilization of the proteins on a solid surface. Cell-free protein array technology attempts to simplify protein microarray construction by bypassing the need to express the proteins in bacteria
Bacteria
Bacteria are a large domain of prokaryotic microorganisms. Typically a few micrometres in length, bacteria have a wide range of shapes, ranging from spheres to rods and spirals...
cells and the subsequent need to purify them. It takes advantage of available cell-free protein synthesis
Cell-free protein synthesis
Cell-free protein synthesis , is the production of protein without the use of living cells.-History:The first elucidation of a codon was done by Marshall Nirenberg and Heinrich J. Matthaei in 1961 at the National Institutes of Health. They used a cell-free system to translate a poly-uracil RNA...
technology which has demonstrated that protein synthesis can occur without an intact cell as long as cell extracts containing the DNA template, transcription
Transcription (genetics)
Transcription is the process of creating a complementary RNA copy of a sequence of DNA. Both RNA and DNA are nucleic acids, which use base pairs of nucleotides as a complementary language that can be converted back and forth from DNA to RNA by the action of the correct enzymes...
and translation raw materials and machinery are provided. Common sources of cell extracts used in cell-free protein array technology include wheat germ, Escherichia coli
Escherichia coli
Escherichia coli is a Gram-negative, rod-shaped bacterium that is commonly found in the lower intestine of warm-blooded organisms . Most E. coli strains are harmless, but some serotypes can cause serious food poisoning in humans, and are occasionally responsible for product recalls...
, and rabbit reticulocyte
Reticulocyte
Reticulocytes are immature red blood cells, typically composing about 1% of the red cells in the human body.Reticulocytes develop and mature in the red bone marrow and then circulate for about a day in the blood stream before developing into mature red blood cells. Like mature red blood cells,...
. Cell extracts from other sources such as hyperthermophile
Hyperthermophile
A hyperthermophile is an organism that thrives in extremely hot environments— from 60 degrees C upwards. An optimal temperature for the existence of hyperthermophiles is above 80°C . Hyperthermophiles are a subset of extremophiles, micro-organisms within the domain Archaea, although some bacteria...
s, hybridoma
Hybridoma
Hybridoma technology is a technology of forming hybrid cell lines by fusing a specific antibody-producing B cell with a myeloma cell that is selected for its ability to grow in tissue culture and for an absence of antibody chain synthesis...
s, Xenopus
Xenopus
Xenopus is a genus of highly aquatic frogs native to Sub-Saharan Africa. There are 19 species in the Xenopus genus...
oocyte
Oocyte
An oocyte, ovocyte, or rarely ocyte, is a female gametocyte or germ cell involved in reproduction. In other words, it is an immature ovum, or egg cell. An oocyte is produced in the ovary during female gametogenesis. The female germ cells produce a primordial germ cell which undergoes a mitotic...
s, insect, mammalian and human cells have also been used.
The target proteins are synthesized in situ
In situ
In situ is a Latin phrase which translated literally as 'In position'. It is used in many different contexts.-Aerospace:In the aerospace industry, equipment on board aircraft must be tested in situ, or in place, to confirm everything functions properly as a system. Individually, each piece may...
on the protein microarray, directly from the DNA template, thus skipping many of the steps in traditional protein microarray production and their accompanying technical limitations. More importantly, the expression of the proteins can be done in parallel, meaning all the proteins can be expressed together in a single reaction. This ability to multiplex protein expression is a major time-saver in the production process.
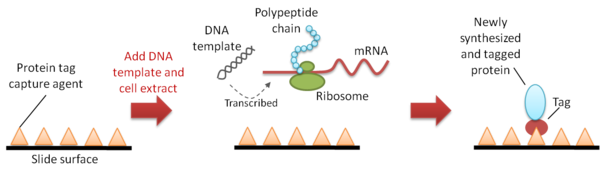
In situ methods
In the in situ method, protein synthesis is carried out on a protein array surface that is pre-coated with a protein-capturing reagent or antibodyAntibody
An antibody, also known as an immunoglobulin, is a large Y-shaped protein used by the immune system to identify and neutralize foreign objects such as bacteria and viruses. The antibody recognizes a unique part of the foreign target, termed an antigen...
. Once the newly synthesized proteins are released from the ribosome
Ribosome
A ribosome is a component of cells that assembles the twenty specific amino acid molecules to form the particular protein molecule determined by the nucleotide sequence of an RNA molecule....
, the tag sequence
Protein tag
Protein tags are peptide sequences genetically grafted onto a recombinant protein. Often these tags are removable by chemical agents or by enzymatic means, such as proteolysis or intein splicing. Tags are attached to proteins for various purposes....
that is also synthesized at the N- or C-terminus of each nascent protein will be bound by the capture reagent or antibody, thus immobilizing the proteins to form an array. Commonly used tags include polyhistidine
Polyhistidine-tag
A polyhistidine-tag is an amino acid motif in proteins that consists of at least five histidine residues, often at the N- or C-terminus of the protein. It is also known as hexa histidine-tag, 6xHis-tag, and by the trademarked name His-tag...
(His)6 and glutathione s-transferase (GST).
Various research groups have developed their own methods, each differing in their approach, but can be summarized into 3 main groups.

Biotinylation
In biochemistry, biotinylation is the process of covalently attaching biotin to a protein, nucleic acid or other molecule. Biotinylation is rapid, specific and is unlikely to perturb the natural function of the molecule due to the small size of biotin...
and is bound to avidin
Avidin
Avidin is a tetrameric biotin-binding protein produced in the oviducts of birds, reptiles and amphibians deposited in the whites of their eggs. In chicken egg white, avidin makes up approximately 0.05% of total protein...
that is pre-coated onto the protein capture surface. Newly synthesized proteins which are tagged with GST are then immobilized next to the template DNA by binding to the adjacent polyclonal anti-GST capture antibody that is also pre-coated onto the capture surface (Figure 1). The main drawback of this method is the extra and tedious preparation steps at the beginning of the process: (1) the cloning
Molecular cloning
Molecular cloning refers to a set of experimental methods in molecular biology that are used to assemble recombinant DNA molecules and to direct their replication within host organisms...
of cDNAs in an expression-ready vector
Expression vector
An expression vector, otherwise known as an expression construct, is generally a plasmid that is used to introduce a specific gene into a target cell. Once the expression vector is inside the cell, the protein that is encoded by the gene is produced by the cellular-transcription and translation...
; and (2) the need to biotinylate the plasmid
Plasmid
In microbiology and genetics, a plasmid is a DNA molecule that is separate from, and can replicate independently of, the chromosomal DNA. They are double-stranded and, in many cases, circular...
DNA but not to interfere with transcription. Moreover, the resulting protein array is not ‘pure’ because the proteins are co-localized with their DNA templates and capture antibodies.


MRNA display
mRNA display is a display technique used for in vitro protein, and/or peptide evolution to create molecules that can bind to a desired target. The process results in translated peptides or proteins that are associated with their mRNA progenitor via a puromycin linkage. The complex then binds to...
technology. PCR DNA is first transcribed to mRNA, and a single-stranded DNA oligonucleotide
Oligonucleotide
An oligonucleotide is a short nucleic acid polymer, typically with fifty or fewer bases. Although they can be formed by bond cleavage of longer segments, they are now more commonly synthesized, in a sequence-specific manner, from individual nucleoside phosphoramidites...
modified with biotin
Biotin
Biotin, also known as Vitamin H or Coenzyme R, is a water-soluble B-complex vitamin discovered by Bateman in 1916. It is composed of a ureido ring fused with a tetrahydrothiophene ring. A valeric acid substituent is attached to one of the carbon atoms of the tetrahydrothiophene ring...
and puromycin
Puromycin
Puromycin is an antibiotic that is a protein synthesis inhibitor by inhibiting translation.-Inhibition of translation:Puromycin is an aminonucleoside antibiotic, derived from the Streptomyces alboniger bacterium, that causes premature chain termination during translation taking place in the...
on each end is then hybridized to the 3’-end of the mRNA. The mRNAs are then arrayed on a slide and immobilized by the binding of biotin to streptavidin
Streptavidin
Streptavidin is a 60000 dalton protein purified from the bacterium Streptomyces avidinii. Streptavidin homo-tetramers have an extraordinarily high affinity for biotin . With a dissociation constant on the order of ≈10-14 mol/L, the binding of biotin to streptavidin is one of the strongest...
that is pre-coated on the slide. Cell extract is then dispensed on the slide for in situ translation to take place. When the ribosome reaches the hybridized oligonucleotide, it stalls and incorporates the puromycin molecule to the nascent polypeptide chain, thereby attaching the newly synthesized protein to the microarray via the DNA oligonucleotide (Figure 3). A pure protein array is obtained after the mRNA is digested with RNase. The protein spots generated by this method are very sharply defined and can be produced at a high density.
Nano-well array format

DNA array to protein array (DAPA)

Advantages
Many of the advantages of cell-free protein array technology address the limitations of cell-based expression system used in traditional methods of protein microarray production.Rapid and cost-effective:
- Avoids DNA cloning (with the exception of NAPPA) and can quickly convert genetic information into functional proteins by using PCR DNA.
- The reduced steps in production and the ability to miniaturize the system saves on reagent consumption and cuts production costs.
Improves protein availability:
- Many proteins, including antibodies, are difficult to express in host cells due to problems with insolubility, disulfideDisulfideIn chemistry, a disulfide usually refers to the structural unit composed of a linked pair of sulfur atoms. Disulfide usually refer to a chemical compound that contains a disulfide bond, such as diphenyl disulfide, C6H5S-SC6H5....
bonds or host cell toxicity. Cell-free protein array makes many of such proteins available for use in protein microarrays.
Enables long term storage
- Unlike DNA, which is a highly stable molecule, proteins are a heterogeneous class of molecules with different stability and physiochemical properties. Maintaining the proteins’ folding and function in an immobilized state over long periods of storage is a major challenge for protein microarrays. Cell-free methods provide the option to quickly obtaining protein microarrays on demand, thus eliminating any problems associated with long-term storage.
Flexible
- Amenable to a range of different templates: PCR products, plasmids and mRNA.
- Additional components can be included during synthesis to adjust the environment for protein folding, disulfide bond formation, modification or protein activity.
Limitation
- Post-translational modification of proteins in proteins generated by cell-free protein synthesis is still limited compared to the traditional methods, and may not be as biologically relevant.
Applications
- Protein interactions: To screen for protein–protein interactions and protein interactions with other molecules such as metaboliteMetaboliteMetabolites are the intermediates and products of metabolism. The term metabolite is usually restricted to small molecules. A primary metabolite is directly involved in normal growth, development, and reproduction. Alcohol is an example of a primary metabolite produced in large-scale by industrial...
s, lipidLipidLipids constitute a broad group of naturally occurring molecules that include fats, waxes, sterols, fat-soluble vitamins , monoglycerides, diglycerides, triglycerides, phospholipids, and others...
s, DNA and small molecules. - Enzyme inhibition assay: For high throughput drug candidate screening and to discover novel enzymeEnzymeEnzymes are proteins that catalyze chemical reactions. In enzymatic reactions, the molecules at the beginning of the process, called substrates, are converted into different molecules, called products. Almost all chemical reactions in a biological cell need enzymes in order to occur at rates...
s for use in biotechnologyBiotechnologyBiotechnology is a field of applied biology that involves the use of living organisms and bioprocesses in engineering, technology, medicine and other fields requiring bioproducts. Biotechnology also utilizes these products for manufacturing purpose...
. - Screening antibody specificity

