Base excision repair
Encyclopedia
Biochemistry
Biochemistry, sometimes called biological chemistry, is the study of chemical processes in living organisms, including, but not limited to, living matter. Biochemistry governs all living organisms and living processes...
and genetics
Genetics
Genetics , a discipline of biology, is the science of genes, heredity, and variation in living organisms....
, base excision repair (BER) is a cellular mechanism that repairs damaged DNA
DNA repair
DNA repair refers to a collection of processes by which a cell identifies and corrects damage to the DNA molecules that encode its genome. In human cells, both normal metabolic activities and environmental factors such as UV light and radiation can cause DNA damage, resulting in as many as 1...
throughout the cell cycle. It is responsible primarily for removing small, non-helix-distorting base lesions from the genome. The related nucleotide excision repair
Nucleotide excision repair
Nucleotide excision repair is a DNA repair mechanism. DNA constantly requires repair due to damage that can occur to bases from a vast variety of sources including chemicals, radiation and other mutagens...
pathway repairs bulky helix-distorting lesions. BER is important for removing damaged bases that could otherwise cause mutation
Mutation
In molecular biology and genetics, mutations are changes in a genomic sequence: the DNA sequence of a cell's genome or the DNA or RNA sequence of a virus. They can be defined as sudden and spontaneous changes in the cell. Mutations are caused by radiation, viruses, transposons and mutagenic...
s by mispairing or lead to breaks in DNA during replication. BER is initiated by DNA glycosylases, which recognize and remove specific damaged or inappropriate bases, forming AP site
AP site
In biochemistry and molecular genetics, an AP site , also known as an abasic site, is a location in DNA that has neither a purine nor a pyrimidine base, either spontaneously or due to DNA damage...
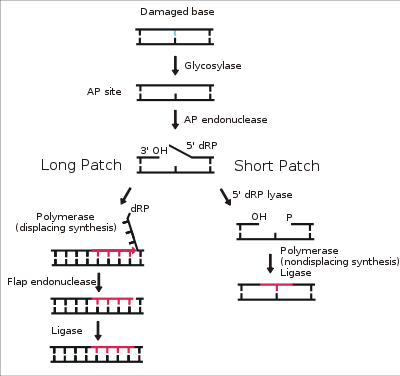
s. These are then cleaved by an AP endonuclease. The resulting single-strand break can then be processed by either short-patch (where a single nucleotide is replaced) or long-patch BER (where 2-10 new nucleotides are synthesized).
Lesions processed by BER
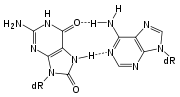
Adenine
Adenine is a nucleobase with a variety of roles in biochemistry including cellular respiration, in the form of both the energy-rich adenosine triphosphate and the cofactors nicotinamide adenine dinucleotide and flavin adenine dinucleotide , and protein synthesis, as a chemical component of DNA...
across from 8-oxoguanine
8-oxoguanine
8-Oxoguanine is one of the most common DNA lesions resulting from reactive oxygen species and can result in a mismatched pairing with Adenine resulting in G to T and C to A substitutions in the genome. In humans, it is primarily repaired by the DNA glycosylase OGG1...
(right) during DNA replication
DNA replication
DNA replication is a biological process that occurs in all living organisms and copies their DNA; it is the basis for biological inheritance. The process starts with one double-stranded DNA molecule and produces two identical copies of the molecule...
causes a G:C base pair to be mutated to T:A. Other examples of base lesions repaired by BER include:
- Oxidized bases: 8-oxoguanine8-oxoguanine8-Oxoguanine is one of the most common DNA lesions resulting from reactive oxygen species and can result in a mismatched pairing with Adenine resulting in G to T and C to A substitutions in the genome. In humans, it is primarily repaired by the DNA glycosylase OGG1...
, 2,6-diamino-4-hydroxy-5-formamidopyrimidine (FapyG, FapyA) - Alkylated bases: 3-methyladenine, 7-methylguanine
- Deaminated bases: hypoxanthineHypoxanthineHypoxanthine is a naturally occurring purine derivative. It is occasionally found as a constituent of nucleic acids where it is present in the anticodon of tRNA in the form of its nucleoside inosine. It has a tautomer known as 6-Hydroxypurine. Hypoxanthine is a necessary additive in certain cell,...
formed from deamination of adenine. XanthineXanthineXanthine , is a purine base found in most human body tissues and fluids and in other organisms. A number of stimulants are derived from xanthine, including caffeine and theobromine....
formed from deamination of guanine. (ThymidineThymidineThymidine is a chemical compound, more precisely a pyrimidine deoxynucleoside. Deoxythymidine is the DNA nucleoside T, which pairs with deoxyadenosine in double-stranded DNA...
products following deamination of 5-methylcytosine5-Methylcytosine5-Methylcytosine is a methylated form of the DNA base cytosine that may be involved in the regulation of gene transcription. When cytosine is methylated, the DNA maintains the same sequence, but the expression of methylated genes can be altered .In the figure on the right, a methyl group, is...
are not repaired) - UracilUracilUracil is one of the four nucleobases in the nucleic acid of RNA that are represented by the letters A, G, C and U. The others are adenine, cytosine, and guanine. In RNA, uracil binds to adenine via two hydrogen bonds. In DNA, the uracil nucleobase is replaced by thymine.Uracil is a common and...
inappropriately incorporated in DNA or formed by deaminationDeaminationDeamination is the removal of an amine group from a molecule. Enzymes which catalyse this reaction are called deaminases.In the human body, deamination takes place primarily in the liver, however glutamate is also deaminated in the kidneys. Deamination is the process by which amino acids are...
of cytosineCytosineCytosine is one of the four main bases found in DNA and RNA, along with adenine, guanine, and thymine . It is a pyrimidine derivative, with a heterocyclic aromatic ring and two substituents attached . The nucleoside of cytosine is cytidine...
In addition to base lesions, the downstream steps of BER are also utilized to repair single-strand breaks.
The choice between long-patch and short-patch repair
The choice between short- and long-patch repair is currently under investigation. Various factors are thought to influence this decision, including the type of lesion, the cell cycle stage, and whether the cell is terminally differentiated or actively dividing. Some lesions, such as oxidized or reduced AP sites, are resistant to pol β lyase activity and, therefore, must be processed by long-patch BER.Pathway preference may differ between organisms, as well. While human cells utilize both short- and long-patch BER, the yeast Saccharomyces cerevisiae
Saccharomyces cerevisiae
Saccharomyces cerevisiae is a species of yeast. It is perhaps the most useful yeast, having been instrumental to baking and brewing since ancient times. It is believed that it was originally isolated from the skin of grapes...
was long thought to lack a short-patch pathway because it does not have homologs of several mammalian short-patch proteins, including pol β, DNA ligase III, XRCC1, and the kinase domain of PNKP
PNKP
Bifunctional polynucleotide phosphatase/kinase is an enzyme that in humans is encoded by the PNKP gene.-Interactions:PNKP has been shown to interact with DNA polymerase beta and XRCC1.-Further reading:...
. The recent discovery that the poly-A polymerase Trf4
TRAMP complex
The TRAMP complex is a multi-protein complex consisting of the RNA helicase Mtr4, a poly polymerase and a zinc knuckle protein...
possesses 5' dRP lyase activity has challenged this view.
DNA glycosylases
AP site
In biochemistry and molecular genetics, an AP site , also known as an abasic site, is a location in DNA that has neither a purine nor a pyrimidine base, either spontaneously or due to DNA damage...
. There are two categories of glycosylases: monofunctional and bifunctional. Monofunctional glycosylases have only glycosylase activity, whereas bifunctional glycosylases also possess AP lyase activity. Therefore, bifunctional glycosylases can convert a base lesion into a single-strand break without the need for an AP endonuclease. β-Elimination of an AP site by a glycosylase-lyase yields a 3' α,β-unsaturated aldehyde adjacent to a 5' phosphate, which differs from the AP endonuclease cleavage product. Some glycosylase-lyases can further perform δ-elimination, which converts the 3' aldehyde to a 3' phosphate. A wide variety of glycosylases have evolved to recognize different damaged bases. Examples of DNA glycosylases include Ogg1
Oxoguanine glycosylase
8-Oxoguanine glycosylase also known as OGG1 is a DNA glycosylase enzyme that, in humans, is encoded by the OGG1 gene. It is involved in base excision repair.- Function :...
, which recognizes 8-oxoguanine, Mag1, which recognizes 3-methyladenine, and UNG
Uracil-DNA glycosylase
Uracil-DNA glycosylase, also known as UNG or UDG, is a human gene though orthologs exist ubiquitously among prokaryotes and eukaryotes and even in some DNA viruses. The first uracil DNA-glycosylase was isolated from Escherichia coli....
, which removes uracil
Uracil
Uracil is one of the four nucleobases in the nucleic acid of RNA that are represented by the letters A, G, C and U. The others are adenine, cytosine, and guanine. In RNA, uracil binds to adenine via two hydrogen bonds. In DNA, the uracil nucleobase is replaced by thymine.Uracil is a common and...
from DNA.
AP endonucleases
The AP endonucleases cleave an AP siteAP site
In biochemistry and molecular genetics, an AP site , also known as an abasic site, is a location in DNA that has neither a purine nor a pyrimidine base, either spontaneously or due to DNA damage...
to yield a 3' hydroxyl adjacent to a 5' deoxyribosephosphate (dRP). AP endonucleases are divided into two families based on their homology to the ancestral bacterial AP endonucleaes endonuclease IV and exonuclease III
Exonuclease III
Exonuclease III is an enzyme that belongs to the exonuclease family. ExoIII catalyzes the stepwise removal of mononucleotides from 3´-hydroxyl termini of duplex DNA...
. Many eukaryotes have members of both families, including the yeast Saccharomyces cerevisiae
Saccharomyces cerevisiae
Saccharomyces cerevisiae is a species of yeast. It is perhaps the most useful yeast, having been instrumental to baking and brewing since ancient times. It is believed that it was originally isolated from the skin of grapes...
, in which Apn1 is the EndoIV homolog and Apn2 is related to ExoIII. In humans, two AP endonucleases, APEX1
APEX1
DNA- lyase is an enzyme that in humans is encoded by the APEX1 gene.-Interactions:APEX1 has been shown to interact with MUTYH, Flap structure-specific endonuclease 1 and XRCC1.-Further reading:...
and APEX2, have been identified. It is a member of the ExoIII family.
End processing enzymes
In order for ligation to occur, a DNA strand break must have a hydroxyl on its 3' end and a phosphate on its 5' end. In humans, polynucleotide kinase-phosphatase (PNKPPNKP
Bifunctional polynucleotide phosphatase/kinase is an enzyme that in humans is encoded by the PNKP gene.-Interactions:PNKP has been shown to interact with DNA polymerase beta and XRCC1.-Further reading:...
) promotes formation of these ends during BER. This protein has a kinase domain, which phosphorylates 5' hydroxyl ends, and a phosphatase domain, which removes phosphates from 3' ends. Together, these activities ready single-strand breaks with damaged termini for ligation. The AP endonucleases also participate in 3' end processing. Besides opening AP sites, they possess 3' phosphodiesterase activity and can remove a variety of 3' lesions including phosphates, phosphoglycolates, and aldehydes. 3'-Processing must occur before DNA synthesis can initiate because DNA polymerases require a 3' hydroxyl to extend from.
DNA polymerases
Pol βDNA polymerase beta
[edit]Polymerase , beta, also known as POLB, is an enzyme that, in humans, is encoded by the POLB gene.- Function :...
is the main human polymerase that catalyzes short-patch BER, with pol λ able to compensate in its absence. These polymerases are members of the Pol X family and typically insert only a single nucleotide. In addition to polymerase activity, these enzymes have a lyase domain that removes the 5' dRP left behind by AP endonuclease cleavage. During long-patch BER, DNA synthesis is thought to be mediated by pol δ and pol ε along with the processivity factor PCNA
PCNA
Proliferating Cell Nuclear Antigen, commonly known as PCNA, is a protein that acts as a processivity factor for DNA polymerase δ in eukaryotic cells. It achieves this processivity by encircling the DNA, thus creating a topological link to the genome...
, the same polymerases that carry out DNA replication
DNA replication
DNA replication is a biological process that occurs in all living organisms and copies their DNA; it is the basis for biological inheritance. The process starts with one double-stranded DNA molecule and produces two identical copies of the molecule...
. These polymerases perform displacing synthesis, meaning that the downstream 5' DNA end is "displaced" to form a flap (see diagram above). Pol β can also perform long-patch displacing synthesis and can, therefore, participate in either BER pathway. Long-patch synthesis typically inserts 2-10 new nucleotides.
Flap endonuclease
FEN1 removes the 5' flap generated during long patch BER. This endonuclease shows a strong preference for a long 5' flap adjacent to a 1-nt 3' flap. The yeast homolog of FEN1 is RAD27. In addition to its role in long-patch BER, FEN1 cleaves flaps with a similar structure during Okazaki fragmentOkazaki fragment
Okazaki fragments are short molecules of single-stranded DNA that are formed on the lagging strand during DNA replication. They are between 1,000 to 2,000 nucleotides long in Escherichia coli and are between 100 to 200 nucleotides long in eukaryotes....
processing, an important step in lagging strand DNA replication
DNA replication
DNA replication is a biological process that occurs in all living organisms and copies their DNA; it is the basis for biological inheritance. The process starts with one double-stranded DNA molecule and produces two identical copies of the molecule...
.
DNA ligase
DNA ligase III along with its cofactor XRCC1XRCC1
XRCC1 is a DNA repair protein.It complexes with DNA ligase III.-Interactions:XRCC1 has been shown to interact with PARP2, DNA polymerase beta, Aprataxin, Oxoguanine glycosylase, PCNA, APEX1, PNKP and PARP1.-Further reading:-External links:...
catalyzes the nick-sealing step in short-patch BER in humans. DNA ligase I ligates the break in long-patch BER.
Links between BER and cancer
Defects in a variety of DNA repair pathways lead to cancer predisposition, and BER appears to follow this pattern. Deletion of BER genes increases the mutation rate in a variety of organisms, predicting that loss of BER could contribute to the development of cancer. Indeed, somatic mutations in Pol β have been found in 30% of human cancers, and some of these mutations lead to transformation when expressed in mouse cells. Mutations in the DNA glycosylase MYHMUTYH
MUTYH is a human gene encoding a DNA glycosylase, MUTYH glycosylase, involved in oxidative DNA damage repair. The enzyme excises adenine bases from the DNA backbone at sites where adenine is inappropriately paired with guanine, cytosine, or 8-oxo-7,8-dihydroguanine, a major oxidatively damaged DNA...
are also known to increase susceptibility to colon cancer.
See also
- DNA repairDNA repairDNA repair refers to a collection of processes by which a cell identifies and corrects damage to the DNA molecules that encode its genome. In human cells, both normal metabolic activities and environmental factors such as UV light and radiation can cause DNA damage, resulting in as many as 1...
- DNA mismatch repairDNA mismatch repairDNA mismatch repair is a system for recognizing and repairing erroneous insertion, deletion and mis-incorporation of bases that can arise during DNA replication and recombination, as well as repairing some forms of DNA damage....
- Nucleotide excision repairNucleotide excision repairNucleotide excision repair is a DNA repair mechanism. DNA constantly requires repair due to damage that can occur to bases from a vast variety of sources including chemicals, radiation and other mutagens...
- Homologous recombinationHomologous recombinationHomologous recombination is a type of genetic recombination in which nucleotide sequences are exchanged between two similar or identical molecules of DNA. It is most widely used by cells to accurately repair harmful breaks that occur on both strands of DNA, known as double-strand breaks...
- Non-homologous end joiningNon-homologous end joiningNon-homologous end joining is a pathway that repairs double-strand breaks in DNA. NHEJ is referred to as "non-homologous" because the break ends are directly ligated without the need for a homologous template, in contrast to homologous recombination, which requires a homologous sequence to guide...

