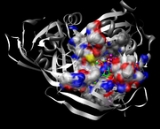
ADP-ribose diphosphatase
Encyclopedia
ADP-ribose diphosphatase is an enzyme
that catalyzes
a hydrolysis reaction in which water nucleophillicly
attacks ADP-ribose to produce AMP
and D-ribose 5-phosphate. Enzyme hydrolysis
occurs by the breakage of a phosphoanhydride bond and is dependent on Mg2+ ions that are held in complex by the enzyme.
.png) The C-terminal domain of ADP-ribose diphosphatase contains the Nudix
The C-terminal domain of ADP-ribose diphosphatase contains the Nudix
sequence, a highly conserved amino acid sequence that is found in over 450 putative proteins in about 90 different species. A part of this sequence known as the Nudix
fold is the catalytic part of the sequence. It is a structurally conserved loop-helix-loop motif that creates a scaffold for metal binding and pyrophosphatase chemistry in the enzyme.
ADP-ribose hydrolases in general act as protective agents against excessive intracellular accumulation of ADP-ribose, as high intracellular levels of ADP-ribose can be damaging to the cell. ADP-ribose diphosphatase, in particular, hydrolyzes ADP-ribose into AMP and D-ribose 5-phosphate, both of which are intermediates of central metabolic pathways and therefore are easily reused.
Other common names for ADP-ribose diphosphatase include ADP-ribose pyrophosphatase and ADPRase. ADP-ribose is commonly referred to as ADPR.
 ADPRase is a dimer
ADPRase is a dimer
of two identical monomers, each of which contain 209 amino acids. The two monomers are folded into two distinct structural domains and with two equivalent catalytic sites. The C-terminal domain consists of the Nudix sequence mentioned above and the N-terminal domain is primarily involved in dimer stabilization. As noted earlier, the Nudix fold is the catalytic part of the enzyme, but it should also be noted that both domains are involved in the active site and they both help with the attachment and coordination of H2O, Mg2+, and the ADP-ribose substrate.
.png) The picture to the right shows the active site of ADPRase in complex with AMPCPR and Mg2+. AMPCPR is a nonhydrolyzable analog of ADP-ribose, and thus functions as an inhibitor of ADPRase. The picture below reveals the first and second Mg2+ ions, which serve to coordinate the attacking water molecule so that it is perfectly in line with the scissile bond (O-P-O bond is 177 degrees) and poised for attack. A glutamate side chain (E162) will deprotonate the water molecule, and then the hydroxide ion will attack nucelophillicly. This occurs when the water molecule is 3.0 angstroms away from the phosphorus molecule it is attacking. By using H2O18, it has been found that the water molecule attacks the adenosyl phosphate. This evidence is a refutation of a previous study that suggested that hydrolysis of ADP-ribose could proceed by nucleophilic attack at either alpha- or beta- phosphate (Gabelli, et al. 2001). After the water molecule attacks, an intermediate is formed, and then ribose 5-phosphate is kicked off as a leaving group. The third active site Mg2+ ion is used to stabilize the negative charge of the phosphate group as it leaves, and is thus situated perfectly between the two phosphates.
The picture to the right shows the active site of ADPRase in complex with AMPCPR and Mg2+. AMPCPR is a nonhydrolyzable analog of ADP-ribose, and thus functions as an inhibitor of ADPRase. The picture below reveals the first and second Mg2+ ions, which serve to coordinate the attacking water molecule so that it is perfectly in line with the scissile bond (O-P-O bond is 177 degrees) and poised for attack. A glutamate side chain (E162) will deprotonate the water molecule, and then the hydroxide ion will attack nucelophillicly. This occurs when the water molecule is 3.0 angstroms away from the phosphorus molecule it is attacking. By using H2O18, it has been found that the water molecule attacks the adenosyl phosphate. This evidence is a refutation of a previous study that suggested that hydrolysis of ADP-ribose could proceed by nucleophilic attack at either alpha- or beta- phosphate (Gabelli, et al. 2001). After the water molecule attacks, an intermediate is formed, and then ribose 5-phosphate is kicked off as a leaving group. The third active site Mg2+ ion is used to stabilize the negative charge of the phosphate group as it leaves, and is thus situated perfectly between the two phosphates.
When the substrate is bound, the structure of the enzyme differs in shape. With the substrate bound, Loop L9 moves 10 angstroms from its original position in the free enzyme, which forms a tighter turn and brings E162 to its catalytic position, meaning this enzyme cycles between an open (free enzyme) and a closed (substrate-metal complex) conformation. This is important because it prevents nonspecific hydrolysis of other nucleotides. ADPRase is highly specific for ADPR, as it has been shown that its Km for ADPR is lower by at least two orders of magnitude than for other sugar nucleotides.
.gif)
. ADP-ribose is a protein-glycating agent, and excess levels of ADP-ribose in the cell can cause non-enzymatic ADP-ribosylation. Non-enzymatic ADP-ribosylation can inactivate protein targets that contain nucleotide-binding sites when the adenylate moiety of ADP-ribose binds to them, and it can also interfere with metabolic regulation that occurs via enzymatic ADP-ribosylation
. For example, actin polymerization is inhibited by non-enzymatic ADP-ribosylation at a Cys
residue. Thus, it is believed that ADPRase functions in general as a house-cleaning enzyme to eliminate potentially deleterious ADP-ribose from the cell.
In the literature, the detoxifying role of ADPRase is directly supported in E. coli cells. But in mammalian cells, there is only an indirect evidence linking ADPRase to a detoxifying role, and this comes from studies of the very specific rat liver ADPRibase-I by cytotoxic agents.
have been solved for this class of enzymes, with PDB
accession codes , , , , , , , , , , , , , , , , , , , , , , , , , and .
Enzyme
Enzymes are proteins that catalyze chemical reactions. In enzymatic reactions, the molecules at the beginning of the process, called substrates, are converted into different molecules, called products. Almost all chemical reactions in a biological cell need enzymes in order to occur at rates...
that catalyzes
Catalysis
Catalysis is the change in rate of a chemical reaction due to the participation of a substance called a catalyst. Unlike other reagents that participate in the chemical reaction, a catalyst is not consumed by the reaction itself. A catalyst may participate in multiple chemical transformations....
a hydrolysis reaction in which water nucleophillicly
Nucleophilic substitution
In organic and inorganic chemistry, nucleophilic substitution is a fundamental class of reactions in which an electron nucleophile selectively bonds with or attacks the positive or partially positive charge of an atom or a group of atoms called the leaving group; the positive or partially positive...
attacks ADP-ribose to produce AMP
Adenosine monophosphate
Adenosine monophosphate , also known as 5'-adenylic acid, is a nucleotide that is used as a monomer in RNA. It is an ester of phosphoric acid and the nucleoside adenosine. AMP consists of a phosphate group, the sugar ribose, and the nucleobase adenine...
and D-ribose 5-phosphate. Enzyme hydrolysis
Hydrolysis
Hydrolysis is a chemical reaction during which molecules of water are split into hydrogen cations and hydroxide anions in the process of a chemical mechanism. It is the type of reaction that is used to break down certain polymers, especially those made by condensation polymerization...
occurs by the breakage of a phosphoanhydride bond and is dependent on Mg2+ ions that are held in complex by the enzyme.
.png)
Nudix family
The Nudix family is a protein family of phosphohydrolases. Using water-mediated catalysis they break a phosphate bond in their substrate to create two products. Nudix stands for Nucleoside Diphosphate linked to X...
sequence, a highly conserved amino acid sequence that is found in over 450 putative proteins in about 90 different species. A part of this sequence known as the Nudix
Nudix family
The Nudix family is a protein family of phosphohydrolases. Using water-mediated catalysis they break a phosphate bond in their substrate to create two products. Nudix stands for Nucleoside Diphosphate linked to X...
fold is the catalytic part of the sequence. It is a structurally conserved loop-helix-loop motif that creates a scaffold for metal binding and pyrophosphatase chemistry in the enzyme.
ADP-ribose hydrolases in general act as protective agents against excessive intracellular accumulation of ADP-ribose, as high intracellular levels of ADP-ribose can be damaging to the cell. ADP-ribose diphosphatase, in particular, hydrolyzes ADP-ribose into AMP and D-ribose 5-phosphate, both of which are intermediates of central metabolic pathways and therefore are easily reused.
Other common names for ADP-ribose diphosphatase include ADP-ribose pyrophosphatase and ADPRase. ADP-ribose is commonly referred to as ADPR.
Structure

Protein dimer
In biochemistry, a dimer is a macromolecular complex formed by two, usually non-covalently bound, macromolecules like proteins or nucleic acids...
of two identical monomers, each of which contain 209 amino acids. The two monomers are folded into two distinct structural domains and with two equivalent catalytic sites. The C-terminal domain consists of the Nudix sequence mentioned above and the N-terminal domain is primarily involved in dimer stabilization. As noted earlier, the Nudix fold is the catalytic part of the enzyme, but it should also be noted that both domains are involved in the active site and they both help with the attachment and coordination of H2O, Mg2+, and the ADP-ribose substrate.
.png)
When the substrate is bound, the structure of the enzyme differs in shape. With the substrate bound, Loop L9 moves 10 angstroms from its original position in the free enzyme, which forms a tighter turn and brings E162 to its catalytic position, meaning this enzyme cycles between an open (free enzyme) and a closed (substrate-metal complex) conformation. This is important because it prevents nonspecific hydrolysis of other nucleotides. ADPRase is highly specific for ADPR, as it has been shown that its Km for ADPR is lower by at least two orders of magnitude than for other sugar nucleotides.
Mechanism
The hydrolysis of ADPR is catalyzed by E162, which improves the nucleophilicity of the water molecule in the active site by deprotonating it. This water is held perfectly in line with the scissile bond by the first and second magnesium ions. The hydroxide ion then attacks the phosphorus atom on the adenosyl phosphate, creating a trigonal bypyramidal intermediate with a negatively charged oxygen attached to the adenosyl phosphate. The double bond is then reformed, effectively discharging ribose 5-phosphate as a leaving group. The third Mg2+ is used to stabilize the negative charge on the leaving group..gif)
Enzyme Functions
ADP-ribose is an intermediate that is produced during the metabolism of NAD+, mono- or poly-unsaturated proteins, and cyclic-ADP riboseAdenosine diphosphate ribose
Adenosine diphosphate ribose is a molecule formed into chains by the enzyme poly ADP ribose polymerase. It binds to and activates the TRPM2 ion channel....
. ADP-ribose is a protein-glycating agent, and excess levels of ADP-ribose in the cell can cause non-enzymatic ADP-ribosylation. Non-enzymatic ADP-ribosylation can inactivate protein targets that contain nucleotide-binding sites when the adenylate moiety of ADP-ribose binds to them, and it can also interfere with metabolic regulation that occurs via enzymatic ADP-ribosylation
ADP-ribosylation
ADP-ribosylation is the addition of one or more ADP-ribose moieties to a protein. These reactions are involved in cell signaling and the control of many cell processes, including DNA repair and apoptosis.-ADP-ribosylation enzymes:...
. For example, actin polymerization is inhibited by non-enzymatic ADP-ribosylation at a Cys
CYS
CYS may refer to:* Cysteine an amino-acid* IATA airport code for Cheyenne Regional Airport.* California Youth Symphony, a San Francisco Bay Area symphony orchestra for young musicians* Count Your Sheep, a webcomic by Adrian Ramos...
residue. Thus, it is believed that ADPRase functions in general as a house-cleaning enzyme to eliminate potentially deleterious ADP-ribose from the cell.
In the literature, the detoxifying role of ADPRase is directly supported in E. coli cells. But in mammalian cells, there is only an indirect evidence linking ADPRase to a detoxifying role, and this comes from studies of the very specific rat liver ADPRibase-I by cytotoxic agents.
PDB Codes
As of late 2007, 26 structuresTertiary structure
In biochemistry and molecular biology, the tertiary structure of a protein or any other macromolecule is its three-dimensional structure, as defined by the atomic coordinates.-Relationship to primary structure:...
have been solved for this class of enzymes, with PDB
Protein Data Bank
The Protein Data Bank is a repository for the 3-D structural data of large biological molecules, such as proteins and nucleic acids....
accession codes , , , , , , , , , , , , , , , , , , , , , , , , , and .

