
UDP-glucuronate decarboxylase
Encyclopedia
In enzymology, an UDP-glucuronate decarboxylase is an enzyme
that catalyzes
the chemical reaction
Hence, this enzyme has one substrate
, UDP-D-glucuronate, and two products
, UDP-D-xylose and CO2
.
This enzyme belongs to the family of lyase
s, specifically the carboxy-lyases, which cleave carbon-carbon bonds. The systematic name of this enzyme class is UDP-D-glucuronate carboxy-lyase (UDP-D-xylose-forming). Other names in common use include uridine-diphosphoglucuronate decarboxylase, and UDP-D-glucuronate carboxy-lyase. This enzyme participates in starch and sucrose metabolism and nucleotide sugars metabolism
. It employs one cofactor
, NAD+.
have been solved for this class of enzymes, with PDB
accession codes and .
Enzyme
Enzymes are proteins that catalyze chemical reactions. In enzymatic reactions, the molecules at the beginning of the process, called substrates, are converted into different molecules, called products. Almost all chemical reactions in a biological cell need enzymes in order to occur at rates...
that catalyzes
Catalysis
Catalysis is the change in rate of a chemical reaction due to the participation of a substance called a catalyst. Unlike other reagents that participate in the chemical reaction, a catalyst is not consumed by the reaction itself. A catalyst may participate in multiple chemical transformations....
the chemical reaction
Chemical reaction
A chemical reaction is a process that leads to the transformation of one set of chemical substances to another. Chemical reactions can be either spontaneous, requiring no input of energy, or non-spontaneous, typically following the input of some type of energy, such as heat, light or electricity...
- UDP-D-glucuronate
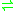 UDP-D-xylose + CO2
UDP-D-xylose + CO2
Hence, this enzyme has one substrate
Substrate (biochemistry)
In biochemistry, a substrate is a molecule upon which an enzyme acts. Enzymes catalyze chemical reactions involving the substrate. In the case of a single substrate, the substrate binds with the enzyme active site, and an enzyme-substrate complex is formed. The substrate is transformed into one or...
, UDP-D-glucuronate, and two products
Product (chemistry)
Product are formed during chemical reactions as reagents are consumed. Products have lower energy than the reagents and are produced during the reaction according to the second law of thermodynamics. The released energy comes from changes in chemical bonds between atoms in reagent molecules and...
, UDP-D-xylose and CO2
Carbon dioxide
Carbon dioxide is a naturally occurring chemical compound composed of two oxygen atoms covalently bonded to a single carbon atom...
.
This enzyme belongs to the family of lyase
Lyase
In biochemistry, a lyase is an enzyme that catalyzes the breaking of various chemical bonds by means other than hydrolysis and oxidation, often forming a new double bond or a new ring structure...
s, specifically the carboxy-lyases, which cleave carbon-carbon bonds. The systematic name of this enzyme class is UDP-D-glucuronate carboxy-lyase (UDP-D-xylose-forming). Other names in common use include uridine-diphosphoglucuronate decarboxylase, and UDP-D-glucuronate carboxy-lyase. This enzyme participates in starch and sucrose metabolism and nucleotide sugars metabolism
Nucleotide sugars metabolism
In nucleotide sugar metabolism a group of biochemicals known as nucleotide sugars act as donors for sugar residues in the glycosylation reactions that produce polysaccharides. They are substrates for glycosyltransferases. The nucleotide sugars are also intermediates in nucleotide sugar...
. It employs one cofactor
Cofactor (biochemistry)
A cofactor is a non-protein chemical compound that is bound to a protein and is required for the protein's biological activity. These proteins are commonly enzymes, and cofactors can be considered "helper molecules" that assist in biochemical transformations....
, NAD+.
Structural studies
As of late 2007, two structuresTertiary structure
In biochemistry and molecular biology, the tertiary structure of a protein or any other macromolecule is its three-dimensional structure, as defined by the atomic coordinates.-Relationship to primary structure:...
have been solved for this class of enzymes, with PDB
Protein Data Bank
The Protein Data Bank is a repository for the 3-D structural data of large biological molecules, such as proteins and nucleic acids....
accession codes and .

