STRING
Encyclopedia
In molecular biology, STRING (Search Tool for the Retrieval of Interacting Genes/Proteins) is a database
and web resource of known and predicted protein-protein interactions.
The STRING database contains information from numerous sources, including experimental data, computational prediction methods and public text collections. It is freely accessible and it is regularly updated. The latest version 9.0 contains information on about 5.2 millions proteins from 1133 species. STRING has been developed by a consortium of academic institutions including CPR
, EMBL, KU
, SIB
, TUD and UZH
.
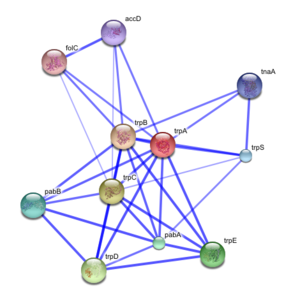
Such networks can be used for filtering and assessing functional genomics data and for providing an intuitive platform for annotating structural, functional and evolutionary properties of proteins.
Exploring the predicted interaction networks can suggest new directions for future experimental research and provide cross-species predictions for efficient interaction mapping.
to use STRING data is available.
Another possibility to access data STRING is to use the application programming interface
(API) by constructing a URL that contain the request.
(iii) interactions transferred from model organisms based on orthology.
All predicted or imported interactions are benchmarked against a common reference of functional partnership as annotated done by KEGG (Kyoto Encyclopedia of Genes and Genomes).
(MINT, HPRD
, BIND, DIP
, BioGRID
, KEGG, Reactome
, IntAct, EcoCyc
, NCI-Nature Pathway Interaction Database
, GO
).
Links are supplied to the originating data of the respective experimental repositories and database resources.
, OMIM, FlyBase
, PubMed
) are parsed to search for statistically relevant co-occurrences of gene names.
Database
A database is an organized collection of data for one or more purposes, usually in digital form. The data are typically organized to model relevant aspects of reality , in a way that supports processes requiring this information...
and web resource of known and predicted protein-protein interactions.
The STRING database contains information from numerous sources, including experimental data, computational prediction methods and public text collections. It is freely accessible and it is regularly updated. The latest version 9.0 contains information on about 5.2 millions proteins from 1133 species. STRING has been developed by a consortium of academic institutions including CPR
Novo Nordisk Foundation Center for Protein Research
The Novo Nordisk Foundation Center for Protein Research , located in the city of Copenhagen, is a subunit of the Faculty of Health Sciences at the University of Copenhagen. It is dedicated to promoting basic and applied discovery research on human proteins of medical relevance...
, EMBL, KU
University of Copenhagen
The University of Copenhagen is the oldest and largest university and research institution in Denmark. Founded in 1479, it has more than 37,000 students, the majority of whom are female , and more than 7,000 employees. The university has several campuses located in and around Copenhagen, with the...
, SIB
Swiss Institute of Bioinformatics
The Swiss Institute of Bioinformatics is an academic not-for-profit foundation which federates bioinformatics activities throughout Switzerland...
, TUD and UZH
University of Zurich
The University of Zurich , located in the city of Zurich, is the largest university in Switzerland, with over 25,000 students. It was founded in 1833 from the existing colleges of theology, law, medicine and a new faculty of philosophy....
.
Usage
Protein-protein interaction networks are an important ingredient for the system-level understanding of cellular processes.Such networks can be used for filtering and assessing functional genomics data and for providing an intuitive platform for annotating structural, functional and evolutionary properties of proteins.
Exploring the predicted interaction networks can suggest new directions for future experimental research and provide cross-species predictions for efficient interaction mapping.

Features
The data is weighted and integrated and a confidence score is calculated for all protein interactions. Results of the various computational predictions can be inspected from different designated views. There are two modes of STRING: Protein-mode and COG-mode. Predicted interactions are propagated to proteins in other organisms for which interaction has been described by inference of orthology. A web interface is available to access the data and to give a fast overview of the proteins and their interactions. A plug-in for cytoscapeCytoscape
Cytoscape is an open source bioinformatics software platform for visualizing molecular interaction networks and integrating with gene expression profiles and other state data. Additional features are available as plugins...
to use STRING data is available.
Another possibility to access data STRING is to use the application programming interface
Application programming interface
An application programming interface is a source code based specification intended to be used as an interface by software components to communicate with each other...
(API) by constructing a URL that contain the request.
Data sources
Like many other database that store protein association knowledge STRING imports data from experimentally derived protein-protein interactions through literature curation. Furthermore, STRING also store computationally predicted interactions from: (i) text mining of scientific texts, (ii) interactions computed from genomic features, and(iii) interactions transferred from model organisms based on orthology.
All predicted or imported interactions are benchmarked against a common reference of functional partnership as annotated done by KEGG (Kyoto Encyclopedia of Genes and Genomes).
Imported data
STRING imports protein association knowledge from databases of physical interaction and databases of curated biological pathway knowledge(MINT, HPRD
HPRD
The Human Protein Reference Database is a protein database accessible through the internet.The HPRD is a result of an international collaborative effort between the in Bangalore, India and the at Johns Hopkins University in Baltimore, USA. HPRD contains manually curated scientific information...
, BIND, DIP
Database of Interacting Proteins
The catalogs experimentally determined interactions between proteins. It combines information from a variety of sources to create a single, consistent set of protein–protein interactions...
, BioGRID
BioGRID
The Biological General Repository for Interaction Datasets is a curated biological database of protein-protein and genetic interactions created in 2003 The Biological General Repository for Interaction Datasets (BioGRID) is a curated biological database of protein-protein and genetic interactions...
, KEGG, Reactome
Reactome
Reactome is a database of biological pathways. There are several Reactomes that concentrate on a specific organism, the largest of these is focused on human biology, but includes pathway steps inferred to exist in humans based on experimental data from model organisms and pathways computationally...
, IntAct, EcoCyc
EcoCyc
EcoCyc is a bioinformatics database for the bacterium Escherichia coli K-12. The EcoCyc project performs literature-based curation of the E. coli genome, and of E. coli transcriptional regulation, transporters, and metabolic pathways....
, NCI-Nature Pathway Interaction Database
NCI-Nature Pathway Interaction Database
The is a free biomedical database of human cellular signaling pathways. The database contains information about the molecular interactions and reactions that take place in cells, with a particular focus on processes that might be relevant to cancer research and treatment. The database was...
, GO
Gene Ontology
The Gene Ontology, or GO, is a major bioinformatics initiative to unify the representation of gene and gene product attributes across all species...
).
Links are supplied to the originating data of the respective experimental repositories and database resources.
Text mining
A large body of scientific texts (SGDSaccharomyces Genome Database
The Saccharomyces Genome Database is a scientific database of the molecular biology and genetics of the yeast Saccharomyces cerevisiae, which is commonly known as baker's or budding yeast....
, OMIM, FlyBase
FlyBase
FlyBase is an online bioinformatics database and the primary repository of genetic and molecular data for the insect family Drosophilidae. For the most extensively studied species and model organism, Drosophila melanogaster, a wide range of data are presented in different formats...
, PubMed
PubMed
PubMed is a free database accessing primarily the MEDLINE database of references and abstracts on life sciences and biomedical topics. The United States National Library of Medicine at the National Institutes of Health maintains the database as part of the Entrez information retrieval system...
) are parsed to search for statistically relevant co-occurrences of gene names.
Predicted data
- Neighborhood: Similar genomic context in different species suggest a similar function of the proteins.
- Fusion-fission events: Proteins that are fused in some genomes are very likely to be functionally linked (as in other genomes where the genes are not fused).
- Occurrence: Proteins that have a similar function or an occurrence in the same metabolic pathway, must be expressed together and have similar phylogenetic profilePhylogenetic profilingPhylogenetic profiling is an important and elegant bioinformatics technique in which the joint presence or joint absence of two traits across a similar distribution of species is used to infer a meaningful biological connection, such as involvement of two different proteins in the same biological...
. - Coexpression: Predicted association between genes based on observed patterns of simultaneous expression of genes.

