
Prokaryotic translation
Encyclopedia
Prokaryotic translation is the process by which messenger RNA
is translated into protein
s in prokaryotes.
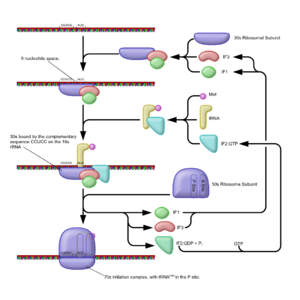 Initiation of translation in prokaryotes involves the assembly of the components of the translation system which are: the two ribosomal
Initiation of translation in prokaryotes involves the assembly of the components of the translation system which are: the two ribosomal
subunits (50S & 30S subunits), the mRNA to be translated, the first (formyl) aminoacyl tRNA (the tRNA charged with the first amino acid
), GTP
(as a source of energy), and three initiation factors
(IF1
, IF2
, and IF3
) which help the assembly of the initiation complex.
The ribosome
has three sites
: the A site, the P site, and the E site. The A site is the point of entry for the aminoacyl tRNA (except for the first aminoacyl tRNA, fMet-tRNAfMet, which enters at the P site). The P site is where the peptidyl tRNA is formed in the ribosome. And the E site which is the exit site of the now uncharged tRNA after it gives its amino acid to the growing peptide chain.
The selection of an initiation site (Usually an AUG codon) depends on the interaction between the 30S RNA and the mRNA template. The 30S subunit binds to the mRNA template at a purine rich region (the Shine Dalgarno sequence) upstream of the AUG initiation codon. The Shine Dalgarno sequence is complementary to a pyrimidine rich region on the 16S rRNA component on the 30S subunit. During the formation of the initiation complex, these complementary nucleotide sequences pair to form a double stranded RNA structure that binds the mRNA to the ribosome in such a way that the initiation codon is placed at the P site.
s to the carboxyl end of the growing chain. The growing protein
exits the ribosome
through the polypeptide exit tunnel in the large subunit.
Elongation starts when the fmet-tRNA enters the P site, causing a conformational change
which opens the A site for the new aminoacyl-tRNA to bind. This binding is facilitated by elongation factor-Tu
(EF-Tu), a small GTPase
. Now the P site contains the beginning of the peptide chain of the protein to be encoded and the A site has the next amino acid to be added to the peptide chain. The growing polypeptide connected to the tRNA in the P site is detached from the tRNA in the P site and a peptide bond
is formed between the last amino acid
s of the polypeptide and the amino acid still attached to the tRNA in the A site. This process, known as peptide bond formation, is catalyzed by a ribozyme (the 23S ribosomal RNA
in the 50S ribosomal subunit). Now, the A site has the newly formed peptide, while the P site has an uncharged tRNA (tRNA with no amino acids). In the final stage of elongation, called translocation, the ribosome moves 3 nucleotides toward the 3' end of the mRNA. Since tRNAs are linked to mRNA by codon-anticodon base-pairing, the tRNAs move relative to the ribosome, taking the nascent polypeptide from the A site to the P site and moving the uncharged tRNA to the E exit site. This process is catalyzed by elongation factor G
(EF-G).
The ribosome continues to translate the remaining codons on the mRNA as more aminoacyl-tRNA bind to the A site, until the ribosome reaches a stop codon on mRNA(UAA, UGA, or UAG).
moves into the A site. These codons are not recognized by any tRNAs. Instead, they are recognized by proteins called release factor
s, namely RF1 (recognizing the UAA and UAG stop codons) or RF2 (recognizing the UAA and UGA stop codons). These factors trigger the hydrolysis
of the ester
bond in peptidyl-tRNA and the release of the newly synthesized protein from the ribosome. A third release factor RF-3 catalyzes the release of RF-1 and RF-2 at the end of the termination process.
and Elongation Factor G (EF-G) function to release mRNA and tRNAs from ribosomes and dissociate the 70S ribosome into the 30S and 50S subunits. IF3 then replaces the deacylated tRNA releasing the mRNA. All translational components are now free for additional rounds of translation.
or polyribosome.
mechanisms to selectively inhibit protein synthesis in bacteria without affecting the host.
Messenger RNA
Messenger RNA is a molecule of RNA encoding a chemical "blueprint" for a protein product. mRNA is transcribed from a DNA template, and carries coding information to the sites of protein synthesis: the ribosomes. Here, the nucleic acid polymer is translated into a polymer of amino acids: a protein...
is translated into protein
Protein
Proteins are biochemical compounds consisting of one or more polypeptides typically folded into a globular or fibrous form, facilitating a biological function. A polypeptide is a single linear polymer chain of amino acids bonded together by peptide bonds between the carboxyl and amino groups of...
s in prokaryotes.
Initiation
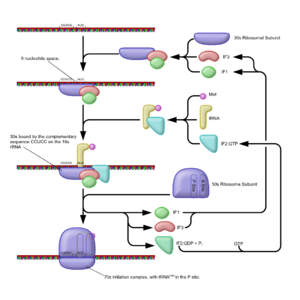
Ribosome
A ribosome is a component of cells that assembles the twenty specific amino acid molecules to form the particular protein molecule determined by the nucleotide sequence of an RNA molecule....
subunits (50S & 30S subunits), the mRNA to be translated, the first (formyl) aminoacyl tRNA (the tRNA charged with the first amino acid
Amino acid
Amino acids are molecules containing an amine group, a carboxylic acid group and a side-chain that varies between different amino acids. The key elements of an amino acid are carbon, hydrogen, oxygen, and nitrogen...
), GTP
Guanosine triphosphate
Guanosine-5'-triphosphate is a purine nucleoside triphosphate. It can act as a substrate for the synthesis of RNA during the transcription process...
(as a source of energy), and three initiation factors
Prokaryotic initiation factors
Prokaryotes require the use of three initiation factors: IF1, IF2, and IF3, for translation.- IF1 :IF1 associates with the 30S ribosomal subunit in the A site and prevents an aminoacyl-tRNA from entering. It modulates IF2 binding to the ribosome by increasing its affinity. It may also prevent the...
(IF1
Prokaryotic initiation factor-1
Prokaryotic initiation factor-1 is a prokaryotic initiation factor.IF1 associates with the 30S ribosomal subunit in the A site and prevents an aminoacyl-tRNA from entering. It modulates IF2 binding to the ribosome by increasing its affinity. It may also prevent the 50S subunit from binding,...
, IF2
Prokaryotic initiation factor-2
Prokaryotic initiation factor-2 is a prokaryotic initiation factor.IF2 binds to an initiator tRNA and controls the entry of that tRNA into the ribosome. IF2, bound to GTP, binds to the 30S P site. After associating with the 30S subunit, fMet-tRNAf binds to the IF2 then IF2 transfers the tRNA into...
, and IF3
Prokaryotic initiation factor-3
Prokaryotic initiation factor-3 is a prokaryotic initiation factor.IF3 is not universally found in all bacterial species but in E. coli it is required for the 30S subunit to bind to the initiation site in mRNA...
) which help the assembly of the initiation complex.
The ribosome
Ribosome
A ribosome is a component of cells that assembles the twenty specific amino acid molecules to form the particular protein molecule determined by the nucleotide sequence of an RNA molecule....
has three sites
Active site
In biology the active site is part of an enzyme where substrates bind and undergo a chemical reaction. The majority of enzymes are proteins but RNA enzymes called ribozymes also exist. The active site of an enzyme is usually found in a cleft or pocket that is lined by amino acid residues that...
: the A site, the P site, and the E site. The A site is the point of entry for the aminoacyl tRNA (except for the first aminoacyl tRNA, fMet-tRNAfMet, which enters at the P site). The P site is where the peptidyl tRNA is formed in the ribosome. And the E site which is the exit site of the now uncharged tRNA after it gives its amino acid to the growing peptide chain.
The selection of an initiation site (Usually an AUG codon) depends on the interaction between the 30S RNA and the mRNA template. The 30S subunit binds to the mRNA template at a purine rich region (the Shine Dalgarno sequence) upstream of the AUG initiation codon. The Shine Dalgarno sequence is complementary to a pyrimidine rich region on the 16S rRNA component on the 30S subunit. During the formation of the initiation complex, these complementary nucleotide sequences pair to form a double stranded RNA structure that binds the mRNA to the ribosome in such a way that the initiation codon is placed at the P site.
Elongation
Elongation of the polypeptide chain involves addition of amino acidAmino acid
Amino acids are molecules containing an amine group, a carboxylic acid group and a side-chain that varies between different amino acids. The key elements of an amino acid are carbon, hydrogen, oxygen, and nitrogen...
s to the carboxyl end of the growing chain. The growing protein
Protein
Proteins are biochemical compounds consisting of one or more polypeptides typically folded into a globular or fibrous form, facilitating a biological function. A polypeptide is a single linear polymer chain of amino acids bonded together by peptide bonds between the carboxyl and amino groups of...
exits the ribosome
Ribosome
A ribosome is a component of cells that assembles the twenty specific amino acid molecules to form the particular protein molecule determined by the nucleotide sequence of an RNA molecule....
through the polypeptide exit tunnel in the large subunit.
Elongation starts when the fmet-tRNA enters the P site, causing a conformational change
Conformational change
A macromolecule is usually flexible and dynamic. It can change its shape in response to changes in its environment or other factors; each possible shape is called a conformation, and a transition between them is called a conformational change...
which opens the A site for the new aminoacyl-tRNA to bind. This binding is facilitated by elongation factor-Tu
Prokaryotic elongation factors
In prokaryotes, three elongation factors are required for translation: EF-Tu, EF-Ts, and EF-G.*EF-Tu mediates the entry of the aminoacyl tRNA into a free site of the ribosome....
(EF-Tu), a small GTPase
GTPase
GTPases are a large family of hydrolase enzymes that can bind and hydrolyze guanosine triphosphate . The GTP binding and hydrolysis takes place in the highly conserved G domain common to all GTPases.-Functions:...
. Now the P site contains the beginning of the peptide chain of the protein to be encoded and the A site has the next amino acid to be added to the peptide chain. The growing polypeptide connected to the tRNA in the P site is detached from the tRNA in the P site and a peptide bond
Peptide bond
This article is about the peptide link found within biological molecules, such as proteins. A similar article for synthetic molecules is being created...
is formed between the last amino acid
Amino acid
Amino acids are molecules containing an amine group, a carboxylic acid group and a side-chain that varies between different amino acids. The key elements of an amino acid are carbon, hydrogen, oxygen, and nitrogen...
s of the polypeptide and the amino acid still attached to the tRNA in the A site. This process, known as peptide bond formation, is catalyzed by a ribozyme (the 23S ribosomal RNA
23S ribosomal RNA
The 23S rRNA is a 2904 nt long component of the large prokaryotic subunit The ribosomal peptidyl transferase activity resides in this rRNA...
in the 50S ribosomal subunit). Now, the A site has the newly formed peptide, while the P site has an uncharged tRNA (tRNA with no amino acids). In the final stage of elongation, called translocation, the ribosome moves 3 nucleotides toward the 3' end of the mRNA. Since tRNAs are linked to mRNA by codon-anticodon base-pairing, the tRNAs move relative to the ribosome, taking the nascent polypeptide from the A site to the P site and moving the uncharged tRNA to the E exit site. This process is catalyzed by elongation factor G
Prokaryotic elongation factors
In prokaryotes, three elongation factors are required for translation: EF-Tu, EF-Ts, and EF-G.*EF-Tu mediates the entry of the aminoacyl tRNA into a free site of the ribosome....
(EF-G).
The ribosome continues to translate the remaining codons on the mRNA as more aminoacyl-tRNA bind to the A site, until the ribosome reaches a stop codon on mRNA(UAA, UGA, or UAG).
Termination
Termination occurs when one of the three termination codonsmoves into the A site. These codons are not recognized by any tRNAs. Instead, they are recognized by proteins called release factor
Release factor
A release factor is a protein that allows for the termination of translation by recognizing the termination codon or stop codon in a mRNA sequence....
s, namely RF1 (recognizing the UAA and UAG stop codons) or RF2 (recognizing the UAA and UGA stop codons). These factors trigger the hydrolysis
Hydrolysis
Hydrolysis is a chemical reaction during which molecules of water are split into hydrogen cations and hydroxide anions in the process of a chemical mechanism. It is the type of reaction that is used to break down certain polymers, especially those made by condensation polymerization...
of the ester
Ester
Esters are chemical compounds derived by reacting an oxoacid with a hydroxyl compound such as an alcohol or phenol. Esters are usually derived from an inorganic acid or organic acid in which at least one -OH group is replaced by an -O-alkyl group, and most commonly from carboxylic acids and...
bond in peptidyl-tRNA and the release of the newly synthesized protein from the ribosome. A third release factor RF-3 catalyzes the release of RF-1 and RF-2 at the end of the termination process.
Recycling
The post-termination complex formed by the end of the termination step consists of mRNA with the termination codon at the A-site, an uncharged tRNA in the P site, and the intact 70S ribosome. Ribosome recycling step is responsible for the disassembly of the post-termination ribosomal complex. Once the nascent protein is released in termination, Ribosome Recycling FactorRibosome Recycling Factor
Ribosome Recycling Factor is a protein found in bacterial cells as well as eukaryotic organelles, specifically mitochondria and chloroplasts. It functions to recycle ribosomes after completion of protein synthesis.- Discovery :...
and Elongation Factor G (EF-G) function to release mRNA and tRNAs from ribosomes and dissociate the 70S ribosome into the 30S and 50S subunits. IF3 then replaces the deacylated tRNA releasing the mRNA. All translational components are now free for additional rounds of translation.
Polysomes
Translation is carried out by more than one ribosome simultaneously. Because of the relatively large size of ribosomes, they can only attach to sites on mRNA 35 nucleotides apart. The complex of one mRNA and a number of ribosomes is called a polysomePolysome
Polyribosomes also known as ergosomes are a cluster of ribosomes, bound to a mRNA molecule, first discovered and characterized by Jonathan Warner, Paul Knopf, and Alex Rich in 1963. Many ribosomes read one mRNA simultaneously, progressing along the mRNA to synthesize the same protein...
or polyribosome.
Effect of antibiotics
Several antibiotics exert their action by targeting the translation process in bacteria. They exploit the differences between prokaryotic and eukaryotic translationEukaryotic translation
Eukaryotic translation is the process by which messenger RNA is translated into proteins in eukaryotes. It consists of initiation, elongation and termination Translation-Initiation:-Cap-dependent initiation:...
mechanisms to selectively inhibit protein synthesis in bacteria without affecting the host.
See also
- Prokaryotic initiation factorsProkaryotic initiation factorsProkaryotes require the use of three initiation factors: IF1, IF2, and IF3, for translation.- IF1 :IF1 associates with the 30S ribosomal subunit in the A site and prevents an aminoacyl-tRNA from entering. It modulates IF2 binding to the ribosome by increasing its affinity. It may also prevent the...
- Prokaryotic elongation factorsProkaryotic elongation factorsIn prokaryotes, three elongation factors are required for translation: EF-Tu, EF-Ts, and EF-G.*EF-Tu mediates the entry of the aminoacyl tRNA into a free site of the ribosome....
- Prokaryotic factors

