Histone H1
Encyclopedia
Histone H1 is one of the five main histone
protein
families which are components of chromatin
in eukaryotic
cells. Though highly conserved
, it is nevertheless the most variable histone in sequence across species.
 Metazoan H1 proteins feature a central globular domain and long C- and short N-terminal tails. H1 is involved with the packing of the "beads on a string" sub-structures into a high order structure, whose details have not yet been solved.
Metazoan H1 proteins feature a central globular domain and long C- and short N-terminal tails. H1 is involved with the packing of the "beads on a string" sub-structures into a high order structure, whose details have not yet been solved.
"bead". Instead, it sits on top of the structure, keeping in place the DNA that has wrapped around the nucleosome. In addition to binding to the nucleosome, the H1 protein binds to the "linker DNA" (approximately 20-80 nucleotides in length) region between nucleosomes, helping stabilize the zig-zagged 30 nm chromatin fiber. Much has been learned about histone H1 from studies on purified chromatin
fibers. Ionic extraction of linker histones from native or reconstituted chromatin promotes its unfolding under hypotonic conditions from fibers of 30 nm width to beads-on-a-string nucleosome arrays.
It is uncertain whether H1 promotes a solenoid-like chromatin fiber, in which exposed linker DNA is shortened, or whether it merely promotes a change in the angle of adjacent nucleosomes, without affecting linker length. Nuclease digestion and DNA footprinting experiments suggest that the globular domain of histone H1 localizes near the nucleosome dyad, where it protects approximately 15-30 base pairs of additional DNA.
In addition, experiments on reconstituted chromatin reveal a characteristic stem motif at the dyad in the presence of H1. Despite gaps in our understanding, a general model has emerged wherein H1’s globular domain closes the nucleosome by crosslinking incoming and outgoing DNA, while the tail binds to linker DNA and neutralizes its negative charge.
Many experiments addressing H1 function have been performed on purified, processed chromatin under low-salt conditions, but H1’s role in vivo is less certain. Cellular studies have shown that overexpression of H1 can cause aberrant nuclear morphology and chromatin structure, and that H1 can serve as both a positive and negative regulator of transcription, depending on the gene. In Xenopus
egg extracts, linker histone depletion causes ~2-fold lengthwise extension of mitotic chromosomes, while overexpression causes chromosomes to hypercompact into an inseparable mass. Complete knockout of H1 in vivo has not been achieved in multicellular organisms due to the existence of multiple isoforms that may be present in several gene clusters, but various linker histone isoforms have been depleted to varying degrees in Tetrahymena, C. elegans, Arabidopsis, fruit fly, and mouse, resulting in various organism-specific defects in nuclear morphology, chromatin structure, DNA methylation, and/or specific gene expression.
It is difficult to understand how such a dynamic protein could be a structural component of chromatin, but it has been suggested that the steady-state equilibrium within the nucleus still strongly favors association between H1 and chromatin, meaning that despite its dynamics, the vast majority of H1 at any given timepoint is chromatin bound.
Cytoplasmic factors appear to be necessary for the dynamic exchange of histone H1 on chromatin, but these have yet to be specifically identified. H1 dynamics may be mediated to some degree by O-glycosylation and phosphorylation. O-glycosylation of H1 may promote chromatin condensation and compaction. Phosphorylation during interphase has been shown to decrease H1 affinity for chromatin and may promote chromatin decondensation and active transcription. However, during mitosis phosphorylation has been shown to increase the affinity of H1 for chromosomes and therefore promote mitotic chromosome condensation (see Mitotic Phosphorylation by CDK1).
. Another isoform is the oocyte/zygotic H1M isoform (also known as B4 or H1foo), found in sea urchins, frogs, mice, and humans, which is replaced in the embryo by somatic isoforms H1A-E, and H10 which resembles H5. Despite having more negative charges than somatic isoforms, H1M binds with higher affinity to mitotic chromosomes in Xenopus
egg extracts.
1 (CDK1) consensus sites, which introduces multiple negative charges. Phosphorylation increases the affinity of H1 for mitotic chromosomes in Xenopus egg extracts and embryos, as determined by fluorescence recovery after photobleaching of wild-type H1 versus non-phosphorylable and phosphomimetic point mutants. These increases in binding affinity appear to offset the cytoplasmic dilution of H1 which occurs in mitotic cells following nuclear envelope
breakdown, illustrating how the cell can regulate H1 to suit specific cellular conditions.
Histone
In biology, histones are highly alkaline proteins found in eukaryotic cell nuclei that package and order the DNA into structural units called nucleosomes. They are the chief protein components of chromatin, acting as spools around which DNA winds, and play a role in gene regulation...
protein
Protein
Proteins are biochemical compounds consisting of one or more polypeptides typically folded into a globular or fibrous form, facilitating a biological function. A polypeptide is a single linear polymer chain of amino acids bonded together by peptide bonds between the carboxyl and amino groups of...
families which are components of chromatin
Chromatin
Chromatin is the combination of DNA and proteins that make up the contents of the nucleus of a cell. The primary functions of chromatin are; to package DNA into a smaller volume to fit in the cell, to strengthen the DNA to allow mitosis and meiosis and prevent DNA damage, and to control gene...
in eukaryotic
Eukaryote
A eukaryote is an organism whose cells contain complex structures enclosed within membranes. Eukaryotes may more formally be referred to as the taxon Eukarya or Eukaryota. The defining membrane-bound structure that sets eukaryotic cells apart from prokaryotic cells is the nucleus, or nuclear...
cells. Though highly conserved
Conserved sequence
In biology, conserved sequences are similar or identical sequences that occur within nucleic acid sequences , protein sequences, protein structures or polymeric carbohydrates across species or within different molecules produced by the same organism...
, it is nevertheless the most variable histone in sequence across species.
Structure

Function
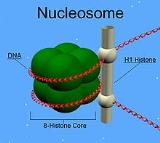
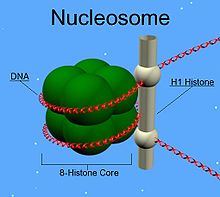
H1 is present in half the amount of the other four histones. This is because unlike the other histones, H1 does not make up the nucleosomeNucleosome
Nucleosomes are the basic unit of DNA packaging in eukaryotes, consisting of a segment of DNA wound around a histone protein core. This structure is often compared to thread wrapped around a spool....
"bead". Instead, it sits on top of the structure, keeping in place the DNA that has wrapped around the nucleosome. In addition to binding to the nucleosome, the H1 protein binds to the "linker DNA" (approximately 20-80 nucleotides in length) region between nucleosomes, helping stabilize the zig-zagged 30 nm chromatin fiber. Much has been learned about histone H1 from studies on purified chromatin
Chromatin
Chromatin is the combination of DNA and proteins that make up the contents of the nucleus of a cell. The primary functions of chromatin are; to package DNA into a smaller volume to fit in the cell, to strengthen the DNA to allow mitosis and meiosis and prevent DNA damage, and to control gene...
fibers. Ionic extraction of linker histones from native or reconstituted chromatin promotes its unfolding under hypotonic conditions from fibers of 30 nm width to beads-on-a-string nucleosome arrays.
It is uncertain whether H1 promotes a solenoid-like chromatin fiber, in which exposed linker DNA is shortened, or whether it merely promotes a change in the angle of adjacent nucleosomes, without affecting linker length. Nuclease digestion and DNA footprinting experiments suggest that the globular domain of histone H1 localizes near the nucleosome dyad, where it protects approximately 15-30 base pairs of additional DNA.
In addition, experiments on reconstituted chromatin reveal a characteristic stem motif at the dyad in the presence of H1. Despite gaps in our understanding, a general model has emerged wherein H1’s globular domain closes the nucleosome by crosslinking incoming and outgoing DNA, while the tail binds to linker DNA and neutralizes its negative charge.
Many experiments addressing H1 function have been performed on purified, processed chromatin under low-salt conditions, but H1’s role in vivo is less certain. Cellular studies have shown that overexpression of H1 can cause aberrant nuclear morphology and chromatin structure, and that H1 can serve as both a positive and negative regulator of transcription, depending on the gene. In Xenopus
Xenopus
Xenopus is a genus of highly aquatic frogs native to Sub-Saharan Africa. There are 19 species in the Xenopus genus...
egg extracts, linker histone depletion causes ~2-fold lengthwise extension of mitotic chromosomes, while overexpression causes chromosomes to hypercompact into an inseparable mass. Complete knockout of H1 in vivo has not been achieved in multicellular organisms due to the existence of multiple isoforms that may be present in several gene clusters, but various linker histone isoforms have been depleted to varying degrees in Tetrahymena, C. elegans, Arabidopsis, fruit fly, and mouse, resulting in various organism-specific defects in nuclear morphology, chromatin structure, DNA methylation, and/or specific gene expression.
Dynamics
A major surprise was the recent discovery from photobleaching experiments that linker histones to be a far more dynamic component of chromatin than core histones, with FRAP studies yielding a t50 of about 1 minute in somatic nuclei.It is difficult to understand how such a dynamic protein could be a structural component of chromatin, but it has been suggested that the steady-state equilibrium within the nucleus still strongly favors association between H1 and chromatin, meaning that despite its dynamics, the vast majority of H1 at any given timepoint is chromatin bound.
Cytoplasmic factors appear to be necessary for the dynamic exchange of histone H1 on chromatin, but these have yet to be specifically identified. H1 dynamics may be mediated to some degree by O-glycosylation and phosphorylation. O-glycosylation of H1 may promote chromatin condensation and compaction. Phosphorylation during interphase has been shown to decrease H1 affinity for chromatin and may promote chromatin decondensation and active transcription. However, during mitosis phosphorylation has been shown to increase the affinity of H1 for chromosomes and therefore promote mitotic chromosome condensation (see Mitotic Phosphorylation by CDK1).
Isoforms
The H1 family in animals includes multiple H1 isoforms that can be expressed in different or overlapping tissues and developmental stages within a single organism. The reason for these multiple isoforms remains unclear, but both their evolutionary conservation from sea urchin to humans as well as significant differences in their amino acid sequences suggest that they are not functionally equivalent. One isoform is histone H5, which is only found in avian erythrocytes, which unlike mammalian erythrocytes, have nucleiCell nucleus
In cell biology, the nucleus is a membrane-enclosed organelle found in eukaryotic cells. It contains most of the cell's genetic material, organized as multiple long linear DNA molecules in complex with a large variety of proteins, such as histones, to form chromosomes. The genes within these...
. Another isoform is the oocyte/zygotic H1M isoform (also known as B4 or H1foo), found in sea urchins, frogs, mice, and humans, which is replaced in the embryo by somatic isoforms H1A-E, and H10 which resembles H5. Despite having more negative charges than somatic isoforms, H1M binds with higher affinity to mitotic chromosomes in Xenopus
Xenopus
Xenopus is a genus of highly aquatic frogs native to Sub-Saharan Africa. There are 19 species in the Xenopus genus...
egg extracts.
Mitotic Phosphorylation by CDK1
At mitosis, somatic H1 isoforms undergo phosphorylation at multiple cyclin-dependent kinaseCyclin-dependent kinase
thumb|350px|Schematic of the cell cycle. outer ring: I=[[Interphase]], M=[[Mitosis]]; inner ring: M=Mitosis; G1=[[G1 phase|Gap phase 1]]; S=[[S phase|Synthesis]]; G2=[[G2 phase|Gap phase 2]]...
1 (CDK1) consensus sites, which introduces multiple negative charges. Phosphorylation increases the affinity of H1 for mitotic chromosomes in Xenopus egg extracts and embryos, as determined by fluorescence recovery after photobleaching of wild-type H1 versus non-phosphorylable and phosphomimetic point mutants. These increases in binding affinity appear to offset the cytoplasmic dilution of H1 which occurs in mitotic cells following nuclear envelope
Nuclear envelope
A nuclear envelope is a double lipid bilayer that encloses the genetic material in eukaryotic cells. The nuclear envelope also serves as the physical barrier, separating the contents of the nucleus from the cytosol...
breakdown, illustrating how the cell can regulate H1 to suit specific cellular conditions.
See also
- nucleosomeNucleosomeNucleosomes are the basic unit of DNA packaging in eukaryotes, consisting of a segment of DNA wound around a histone protein core. This structure is often compared to thread wrapped around a spool....
- histoneHistoneIn biology, histones are highly alkaline proteins found in eukaryotic cell nuclei that package and order the DNA into structural units called nucleosomes. They are the chief protein components of chromatin, acting as spools around which DNA winds, and play a role in gene regulation...
- chromatinChromatinChromatin is the combination of DNA and proteins that make up the contents of the nucleus of a cell. The primary functions of chromatin are; to package DNA into a smaller volume to fit in the cell, to strengthen the DNA to allow mitosis and meiosis and prevent DNA damage, and to control gene...
. - Other histoneHistoneIn biology, histones are highly alkaline proteins found in eukaryotic cell nuclei that package and order the DNA into structural units called nucleosomes. They are the chief protein components of chromatin, acting as spools around which DNA winds, and play a role in gene regulation...
proteins involved in chromatin: - H2AHistone H2AHistone H2A is one of the 5 main histone proteins involved in the structure of chromatin in eukaryotic cells. Featuring a main globular domain and a long N terminal tail, H2A is involved with the structure of the nucleosomes of the 'beads on a string' structure.Other histone proteins...
- H2BHistone H2BHistone H2B is one of the 5 main histone proteins involved in the structure of chromatin in eukaryotic cells. Featuring a main globular domain and a long N terminal tail H2B is involved with the structure of the nucleosomes of the 'beads on a string' structure.See nucleosome, histone and...
- H3Histone H3Histone H3 is one of the five main histone proteins involved in the structure of chromatin in eukaryotic cells. Featuring a main globular domain and a long N-terminal tail, H3 is involved with the structure of the nucleosomes of the 'beads on a string' structure...
- H4Histone H4Histone H4 is one of the 5 main histone proteins involved in the structure of chromatin in eukaryotic cells. Featuring a main globular domain and a long N terminal tail, H4 is a structural component of the nucleosome, and is subject to covalent modification, including acetylation and methylation,...

