
Generalized tree alignment
Encyclopedia
In computational phylogenetics
, generalized tree alignment is the problem of producing a multiple sequence alignment
and a phylogenetic tree
on a set of sequences simultaneously, as opposed to separately.
Formally, Generalized tree alignment is the following optimization problem.
Input: A set and an edit distance function
and an edit distance function 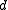 between sequences,
between sequences,
Output: A tree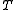 leaf-labeled by
leaf-labeled by 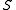 and labeled with sequences at the internal nodes, such that
and labeled with sequences at the internal nodes, such that 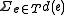 is minimized, where
is minimized, where 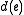 is the edit distance between the endpoints of
is the edit distance between the endpoints of  .
.
Note that this is in contrast to tree alignment
, where the tree is provided as input.
Phylogenetics
In biology, phylogenetics is the study of evolutionary relatedness among groups of organisms , which is discovered through molecular sequencing data and morphological data matrices...
, generalized tree alignment is the problem of producing a multiple sequence alignment
Multiple sequence alignment
A multiple sequence alignment is a sequence alignment of three or more biological sequences, generally protein, DNA, or RNA. In many cases, the input set of query sequences are assumed to have an evolutionary relationship by which they share a lineage and are descended from a common ancestor...
and a phylogenetic tree
Phylogenetic tree
A phylogenetic tree or evolutionary tree is a branching diagram or "tree" showing the inferred evolutionary relationships among various biological species or other entities based upon similarities and differences in their physical and/or genetic characteristics...
on a set of sequences simultaneously, as opposed to separately.
Formally, Generalized tree alignment is the following optimization problem.
Input: A set
 and an edit distance function
and an edit distance function 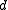 between sequences,
between sequences,Output: A tree
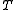 leaf-labeled by
leaf-labeled by 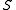 and labeled with sequences at the internal nodes, such that
and labeled with sequences at the internal nodes, such that 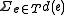 is minimized, where
is minimized, where 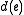 is the edit distance between the endpoints of
is the edit distance between the endpoints of  .
.Note that this is in contrast to tree alignment
Tree alignment
In computational phylogenetics, tree alignment is the problem of producing a multiple sequence alignment on a set of sequences over a fixed tree.Formally, tree alignment is the following optimization problem....
, where the tree is provided as input.

