Coronaviridae
Encyclopedia
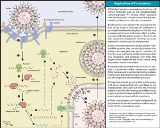
Coronaviruses are enveloped, single-stranded, positive-sense RNA
viruses (27-31kb) with club-shaped surface about 120-160 nm in diameter that resemble a “corona”.
consists of two viral-specified structural glycoproteins S and M. Glycoprotein
S is located on the outer membrane of the envelope and is responsible for the club-shaped projections. On the other hand, glycoprotein M is a transmembrane molecule and is located on the inner part of the envelope. Another important structural protein
is the phosphoprotein
N, which is responsible for the helical symmetry of the nucleocapsid that encloses the genomic RNA.
es, including the following subfamilies:
In April of 2008, the following proposals were ratified by the ICTV:
In July of 2009, the following proposals were ratified by the ICTV:
of the respiratory tract known as Severe Acute Respiratory Syndrome
(SARS). They can also cause enteric infections in very young infants and, in rare situations, neurological syndromes.
 Coronaviruses have single-stranded, positive-sense RNA
Coronaviruses have single-stranded, positive-sense RNA
genome
s of about 30 kilobases, by far the largest non-segmented RNA virus genomes currently known. The key functions required for coronavirus RNA synthesis are encoded by the viral replicase gene. The gene comprises more than 20,000 nucleotide
s and encodes two replicase polyproteins, pp1a and pp1ab, that are proteolytically processed by viral protease
s. Over the past years, it has become clear that the unique size of the coronavirus genome and the special mechanism that coronaviruses (and several other nidoviruses) have evolved to produce an extensive set of subgenome-length RNA
s is linked to the production of a number of nonstructural proteins (nsps) that is unprecedented among RNA viruses. Many of these replicase cleavage products are in fact multidomain proteins themselves, thus further increasing the complexity of protein functions and interactions. Structural studies suggest that several nsps, following their release from larger precursor molecules, form dimer
s or even multimers. The various pp1a/pp1ab precursors and processing products are thought to assemble into large, membrane-associated complexes that, in a temporally coordinated manner, catalyze the reactions involved in RNA replication and transcription
and, it is presumed, serve yet other functions in the viral life cycle.
Coronaviruses also exhibit ribosomal frameshifting and polymerase stuttering
as part of their complex replicative cycle
.
RNA
Ribonucleic acid , or RNA, is one of the three major macromolecules that are essential for all known forms of life....
viruses (27-31kb) with club-shaped surface about 120-160 nm in diameter that resemble a “corona”.
Virology
The 5' and 3' ends of the genome have a cap and poly (A) tract, respectively. The envelopeEnvelope
An envelope is a common packaging item, usually made of thin flat material. It is designed to contain a flat object, such as a letter or card....
consists of two viral-specified structural glycoproteins S and M. Glycoprotein
Glycoprotein
Glycoproteins are proteins that contain oligosaccharide chains covalently attached to polypeptide side-chains. The carbohydrate is attached to the protein in a cotranslational or posttranslational modification. This process is known as glycosylation. In proteins that have segments extending...
S is located on the outer membrane of the envelope and is responsible for the club-shaped projections. On the other hand, glycoprotein M is a transmembrane molecule and is located on the inner part of the envelope. Another important structural protein
Protein
Proteins are biochemical compounds consisting of one or more polypeptides typically folded into a globular or fibrous form, facilitating a biological function. A polypeptide is a single linear polymer chain of amino acids bonded together by peptide bonds between the carboxyl and amino groups of...
is the phosphoprotein
Phosphoprotein
Phosphoproteins are proteins that are physically bonded to a substance containing phosphoric acid . This category of organic molecules includes Fc receptors, Ulks, Calcineurins, K chips, and urocortins....
N, which is responsible for the helical symmetry of the nucleocapsid that encloses the genomic RNA.
Taxonomy
The Coronaviridae are a family of virusVirus
A virus is a small infectious agent that can replicate only inside the living cells of organisms. Viruses infect all types of organisms, from animals and plants to bacteria and archaea...
es, including the following subfamilies:
- Subfamily: Coronavirinae
- Genera: Alphacoronavirus, Betacoronavirus, Gammacoronavirus
- Genera: Alphacoronavirus, Betacoronavirus, Gammacoronavirus
- Subfamily: Torovirinae
- Genera: Bafinivirus, TorovirusTorovirusTorovirus is a genus of viruses within the Coronaviridae family that primarily infect vertebrates. They cause gastroenteritis in mammals, including humans but rarely. Torovirus particles share characteristics with other members of the coronavirus family; they are round, pleomorphic, enveloped...
- Genera: Bafinivirus, Torovirus
In April of 2008, the following proposals were ratified by the ICTV:
- 2005.260V.04 To create the following species in the genus Coronavirus in the family Coronaviridae, named Goose coronavirus, Pigeon coronavirus, Duck coronavirus.
- 2006.009V.04 To create a species in the genus Coronavirus in the family Coronaviridae, named Human coronavirus NL63.
- 2006.010V.04 To create a species in the genus Coronavirus in the family Coronaviridae, named Human coronavirus HKU1.
- 2006.011V.04 To create a species in the genus Coronavirus in the family Coronaviridae, namedEquine coronavirus.
In July of 2009, the following proposals were ratified by the ICTV:
- 2008.085-122V.A.v3.Coronaviridae
- 2008.085V Create a new subfamily in the family Coronaviridae, order Nidovirales
- 2008.086V Name the new subfamily Coronavirinae
- 2008.087V Create a new genus in the proposed subfamily Coronavirinae
- 2008.088V Name the new genus Alphacoronavirus
- 2008.089V Assign three existing species (Human coronavirus 229E, Human coronavirus NL63, Porcine epidemic diarrhea virus) and five new species proposed in 2008.091-095V.01 to the proposed new genus Alphacoronavirus
- 2008.090V Designate proposed species Alphacoronavirus 1 as type species of the genus Alphacoronavirus
- 2008.091V Create new species named Alphacoronavirus 1 in the new genus
- 2008.092V Create new species named Rhinolophus bat coronavirus HKU2 in the new genus
- 2008.093V Create new species named Scotophilus bat coronavirus 512 in the new genus
- 2008.094V Create new species named Miniopterus bat coronavirus 1 in the new genus
- 2008.095V Create new species named Miniopterus bat coronavirus HKU8 in the new genus
- 2008.096V Create a new genus in the proposed subfamily Coronavirinae
- 2008.097V Name the new genus Betacoronavirus
- 2008.098V Assign the existing species Human coronavirus HKU1 and six new species proposed in
- 2008.100-105V.01 to the proposed genus Betacoronavirus
- 2008.099V Designate proposed species Murine coronavirus as type species of the genus Betacoronavirus
- 2008.108V Assign the two species proposed in 2008.110,111V.01 to the new genus
- 2008.109V Designate proposed species Avian coronavirus as type species of the new genus
- 2008.110V Create species named Avian coronavirus in the new genus
- 2008.111V Create species named Beluga whale coronavirus SW1 in the new genus
- 2008.112V Create a new subfamily in the family Coronaviridae, order Nidovirales
- 2008.113V Name the new subfamily Torovirinae
- 2008.114V Create a new genus in the subfamily Torovirinae
- 2008.115V Name the new genus Bafinivirus
- 2008.116V Assign the species White breamVirus (proposed in 2008.118V.01) to the new genus
- 2008.117V Designate species White bream virus as type species in the new genus
- 2008.118V Create species named White bream virus in the new genus
- 2008.119V Remove the genus Torovirus from the family Coronaviridae
- 2008.120V Reassign the genus Torovirus to the subfamily Torovirinae
- 2008.121V.U Remove (abolish) 18 species (Human enteric coronavirus, Human coronavirus OC43, Bovine coronavirus, Porcine hemagglutinating encephalomyelitis virus, Equine coronavirus, Murine hepatitis virus, Puffinosis coronavirus, Rat coronavirus, Transmissible gastroenteritis virus, Canine coronavirus, Feline coronavirus, Infectious bronchitis virus, Duck coronavirus, Goose coronavirus, Pheasant coronavirus, Pigeon coronavirus, Turkey coronavirus, Severe acute respiratory syndrome coronavirus) from the genus Coronavirus
- 2008.122V.U Reassign species Human coronavirus 229E, Human coronavirus NL63 and Porcine epidemic diarrhea virus to the new genus Alphacoronavirus and Human coronavirus HKU1 to the new genus Betacoronavirus
Transmission
Coronaviruses are transmitted by faecal-oral route or by aerosols of respiratory secretions.Pathogenesis
Coronaviruses infect a wide range of mammals and birds and occur worldwide. Although most diseases are mild, sometimes they can cause more severe situations in humans, such as, for example, the infectionInfection
An infection is the colonization of a host organism by parasite species. Infecting parasites seek to use the host's resources to reproduce, often resulting in disease...
of the respiratory tract known as Severe Acute Respiratory Syndrome
Severe acute respiratory syndrome
Severe Acute Respiratory Syndrome is a respiratory disease in humans which is caused by the SARS coronavirus . Between November 2002 and July 2003 an outbreak of SARS in Hong Kong nearly became a pandemic, with 8,422 cases and 916 deaths worldwide according to the WHO...
(SARS). They can also cause enteric infections in very young infants and, in rare situations, neurological syndromes.
SARS
Human infection by SARS coronavirus appears to be limited to the respiratory tract where infection of susceptible cells leads to damage to the pneumocytes resulting in a histological picture of diffuse alveolar damage and a clinical picture of adult respiratory distress syndrome. Diarrhea is also present but there is limited evidence of damage to the intestinal epithelium. The damage to the respiratory tree appears limited to the lower respiratory tract and there is evidence that the immune response plays a part in the outcome of patients with SARS.Binding and Entry
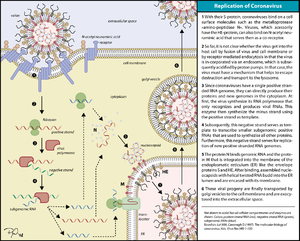
Coronaviruses bind to host cells primarily through interactions between viral spike glycoproteins and specific host cell surface glycoproteins. Some coronaviruses also bind to sialic acids on glycoproteins and glycolipids via their spike and/or hemaglutinin esterase glycoproteins. The interactions between coronaviruses and host cell receptors are critical determinants of species-specificity, tissue tropism, and virulence.Replication

RNA
Ribonucleic acid , or RNA, is one of the three major macromolecules that are essential for all known forms of life....
genome
Genome
In modern molecular biology and genetics, the genome is the entirety of an organism's hereditary information. It is encoded either in DNA or, for many types of virus, in RNA. The genome includes both the genes and the non-coding sequences of the DNA/RNA....
s of about 30 kilobases, by far the largest non-segmented RNA virus genomes currently known. The key functions required for coronavirus RNA synthesis are encoded by the viral replicase gene. The gene comprises more than 20,000 nucleotide
Nucleotide
Nucleotides are molecules that, when joined together, make up the structural units of RNA and DNA. In addition, nucleotides participate in cellular signaling , and are incorporated into important cofactors of enzymatic reactions...
s and encodes two replicase polyproteins, pp1a and pp1ab, that are proteolytically processed by viral protease
Protease
A protease is any enzyme that conducts proteolysis, that is, begins protein catabolism by hydrolysis of the peptide bonds that link amino acids together in the polypeptide chain forming the protein....
s. Over the past years, it has become clear that the unique size of the coronavirus genome and the special mechanism that coronaviruses (and several other nidoviruses) have evolved to produce an extensive set of subgenome-length RNA
Subgenomic mRNA
Subgenomic mRNAs are essentially smaller sections of the original transcribed template strand. During transcription, the original template strand is usually read from the 3' to the 5' end from beginning to end...
s is linked to the production of a number of nonstructural proteins (nsps) that is unprecedented among RNA viruses. Many of these replicase cleavage products are in fact multidomain proteins themselves, thus further increasing the complexity of protein functions and interactions. Structural studies suggest that several nsps, following their release from larger precursor molecules, form dimer
Protein dimer
In biochemistry, a dimer is a macromolecular complex formed by two, usually non-covalently bound, macromolecules like proteins or nucleic acids...
s or even multimers. The various pp1a/pp1ab precursors and processing products are thought to assemble into large, membrane-associated complexes that, in a temporally coordinated manner, catalyze the reactions involved in RNA replication and transcription
Transcription (genetics)
Transcription is the process of creating a complementary RNA copy of a sequence of DNA. Both RNA and DNA are nucleic acids, which use base pairs of nucleotides as a complementary language that can be converted back and forth from DNA to RNA by the action of the correct enzymes...
and, it is presumed, serve yet other functions in the viral life cycle.
Coronaviruses also exhibit ribosomal frameshifting and polymerase stuttering
Polymerase stuttering
Polymerase stuttering is the process by which a polymerase transcribes a nucleotide several times without progressing further on the mRNA chain. It is often used in addition of poly A tails or capping mRNA chains by less complex organisms such as viruses....
as part of their complex replicative cycle
Viral life cycle
Viruses are similar to living organisms, however there are differences. One of the ways a virus can be seen as living is that a virus needs to replicate and create progeny. However, unlike other organisms, a virus cannot survive on its own. It is only active when replicating within a host, using a...
.

