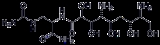
Zwittermicin A
Encyclopedia
Zwittermicin A is an antibiotic that has been identified from the bacterium Bacillus cereus
UW85.. It is a molecule of interest to agricultural industry because it has the potential to suppress plant disease due to its broad spectrum activity against certain gram positive and gram negative eukaryotic micro-organisms. The molecule is also of interest from a metabolic perspective because it represents a new structural class of antibiotic and suggests a crossover between polyketide
and non-ribosomal peptide biosynthetic pathways. Zwittermicin A is linear aminopolyol.
is a hybrid of polyketide and non-ribosomal peptide synthetic pathways. Most likely, all of the synthases are located on one megasynthase much like a type I ketosynthase. Based on mutant studies, the biosynthetic cluster involved in zwittermicin production have been identified and the pathway proposed. The gene
s responsible for the production of zwittermicin A are located on a 16 kb cluster containing nine orfs and a self resistant gene zmaR, a gene
that encodes an acylation
enzyme
that deactivate zwittermicin A. The hybrid synthase
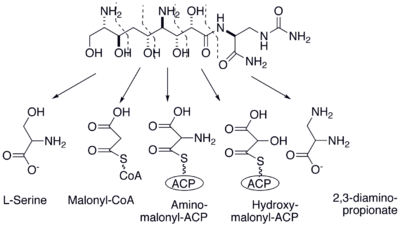
used in zwittermicin A production utilizes modified extender units such as hydroxymalonyl-ACP, aminomalonyl-ACP and 2,3 diamino propionate. Therefore, many of the genes in the biosynthetic cluster encode for enzyme
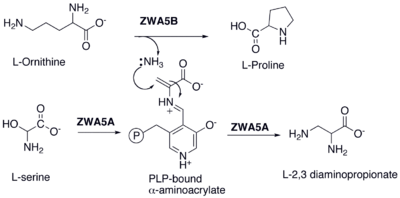
s responsible for the synthesis of these extender units used in the hybrid synthase. For example, orf5 encodes ZWA5A, an enzyme that is responsible for the PLP mediated amination that converts L-serine to 2,3 diaminopropionate. It has also been shown that orf5, orf7, orf4 and orf6 participate in the biosynthesis of aminomalonyl-ACP and orf3, orf2 and orf1 synthesize hydroxymalonyl-ACP.
 Gene organization of the Zwittermicin A biosynthetic cluster.
Gene organization of the Zwittermicin A biosynthetic cluster.
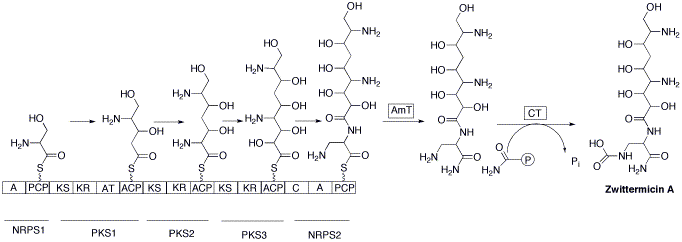
Genes encoding for the seven component hybrid synthase responsible for the assembly of the backbone is likely located on the largest gene, orf8. Assembly begins by the activation of a serine residue. This is done by tethering the amino acid to a peptidal carrier protein via a non-ribosomal peptide synthetase. Subsequently elongation of an activated malonyl unit covalently attached to an acyl carrier protein by a ketosynthase occurs giving the five carbon unit. The next two elongation steps proceed in a similar manner using aminomalonyl and hydroxymalonyl units from a second and third ketosynthase. Finally, condensation of 2,3-diaminopropionate with the carried molecule by a second nonribosomalpeptide synthase produces the zwittermicin A backbone. Attack of ammonia via an amidotransferase enzyme releases the carrier protein. The last step involves a carbomyltransferase enzyme that carbamolates the released molecule giving the final product.
Bacillus cereus
Bacillus cereus is an endemic, soil-dwelling, Gram-positive, rod-shaped, beta hemolytic bacterium. Some strains are harmful to humans and cause foodborne illness, while other strains can be beneficial as probiotics for animals...
UW85.. It is a molecule of interest to agricultural industry because it has the potential to suppress plant disease due to its broad spectrum activity against certain gram positive and gram negative eukaryotic micro-organisms. The molecule is also of interest from a metabolic perspective because it represents a new structural class of antibiotic and suggests a crossover between polyketide
Polyketide
Polyketides are secondary metabolites from bacteria, fungi, plants, and animals. Polyketides are usually biosynthesized through the decarboxylative condensation of malonyl-CoA derived extender units in a similar process to fatty acid synthesis...
and non-ribosomal peptide biosynthetic pathways. Zwittermicin A is linear aminopolyol.
Biosynthesis
Zwittermycin A biosynthesisBiosynthesis
Biosynthesis is an enzyme-catalyzed process in cells of living organisms by which substrates are converted to more complex products. The biosynthesis process often consists of several enzymatic steps in which the product of one step is used as substrate in the following step...
is a hybrid of polyketide and non-ribosomal peptide synthetic pathways. Most likely, all of the synthases are located on one megasynthase much like a type I ketosynthase. Based on mutant studies, the biosynthetic cluster involved in zwittermicin production have been identified and the pathway proposed. The gene
Gene
A gene is a molecular unit of heredity of a living organism. It is a name given to some stretches of DNA and RNA that code for a type of protein or for an RNA chain that has a function in the organism. Living beings depend on genes, as they specify all proteins and functional RNA chains...
s responsible for the production of zwittermicin A are located on a 16 kb cluster containing nine orfs and a self resistant gene zmaR, a gene
Gene
A gene is a molecular unit of heredity of a living organism. It is a name given to some stretches of DNA and RNA that code for a type of protein or for an RNA chain that has a function in the organism. Living beings depend on genes, as they specify all proteins and functional RNA chains...
that encodes an acylation
Acylation
In chemistry, acylation is the process of adding an acyl group to a compound. The compound providing the acyl group is called the acylating agent....
enzyme
Enzyme
Enzymes are proteins that catalyze chemical reactions. In enzymatic reactions, the molecules at the beginning of the process, called substrates, are converted into different molecules, called products. Almost all chemical reactions in a biological cell need enzymes in order to occur at rates...
that deactivate zwittermicin A. The hybrid synthase
Synthase
In biochemistry, a synthase is an enzyme that catalyses a synthesis process.Following the EC number classification, they belong to the group of ligases, with lyases catalysing the reverse reaction....
used in zwittermicin A production utilizes modified extender units such as hydroxymalonyl-ACP, aminomalonyl-ACP and 2,3 diamino propionate. Therefore, many of the genes in the biosynthetic cluster encode for enzyme
Enzyme
Enzymes are proteins that catalyze chemical reactions. In enzymatic reactions, the molecules at the beginning of the process, called substrates, are converted into different molecules, called products. Almost all chemical reactions in a biological cell need enzymes in order to occur at rates...
s responsible for the synthesis of these extender units used in the hybrid synthase. For example, orf5 encodes ZWA5A, an enzyme that is responsible for the PLP mediated amination that converts L-serine to 2,3 diaminopropionate. It has also been shown that orf5, orf7, orf4 and orf6 participate in the biosynthesis of aminomalonyl-ACP and orf3, orf2 and orf1 synthesize hydroxymalonyl-ACP.



Genes encoding for the seven component hybrid synthase responsible for the assembly of the backbone is likely located on the largest gene, orf8. Assembly begins by the activation of a serine residue. This is done by tethering the amino acid to a peptidal carrier protein via a non-ribosomal peptide synthetase. Subsequently elongation of an activated malonyl unit covalently attached to an acyl carrier protein by a ketosynthase occurs giving the five carbon unit. The next two elongation steps proceed in a similar manner using aminomalonyl and hydroxymalonyl units from a second and third ketosynthase. Finally, condensation of 2,3-diaminopropionate with the carried molecule by a second nonribosomalpeptide synthase produces the zwittermicin A backbone. Attack of ammonia via an amidotransferase enzyme releases the carrier protein. The last step involves a carbomyltransferase enzyme that carbamolates the released molecule giving the final product.


