
Template Modeling Score (bioinformatics)
Encyclopedia
The Template Modeling Score or TM-score is a measure of similarity between two protein structure
s with different tertiary structure
s. The TM-score is intended as a more accurate measure of the quality of full-length protein structures than the often used RMSD and GDT
measures. The TM-score indicates the difference between two structures by a score between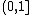 , where 1 indicates a perfect match between two structures. Generally scores below 0.20 corresponds to randomly chosen unrelated proteins whereas structures with a score higher than 0.5 assume roughly the same fold.
, where 1 indicates a perfect match between two structures. Generally scores below 0.20 corresponds to randomly chosen unrelated proteins whereas structures with a score higher than 0.5 assume roughly the same fold.
A quantitative study
shows that proteins of TM-score = 0.5 have a posterior probability
of 37% in the same CATH
Topology family and of 13% in the same SCOP
Fold family. The probabilities increase rapidly when TM-score >0.5. The TM-score is designed to be independent of protein lengths.
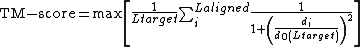
where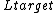 and
and 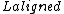 are the lengths of the target protein and the aligned region respectively.
are the lengths of the target protein and the aligned region respectively. 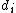 is the distance between the
is the distance between the 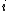 th pair of residues and
th pair of residues and
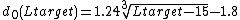
is a distance scale that normalizes distances.
Protein structure
Proteins are an important class of biological macromolecules present in all organisms. Proteins are polymers of amino acids. Classified by their physical size, proteins are nanoparticles . Each protein polymer – also known as a polypeptide – consists of a sequence formed from 20 possible L-α-amino...
s with different tertiary structure
Tertiary structure
In biochemistry and molecular biology, the tertiary structure of a protein or any other macromolecule is its three-dimensional structure, as defined by the atomic coordinates.-Relationship to primary structure:...
s. The TM-score is intended as a more accurate measure of the quality of full-length protein structures than the often used RMSD and GDT
Global distance test
The global distance test or GDT is a measure of similarity between two protein structures with identical amino acid sequences but different tertiary structures...
measures. The TM-score indicates the difference between two structures by a score between
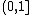 , where 1 indicates a perfect match between two structures. Generally scores below 0.20 corresponds to randomly chosen unrelated proteins whereas structures with a score higher than 0.5 assume roughly the same fold.
, where 1 indicates a perfect match between two structures. Generally scores below 0.20 corresponds to randomly chosen unrelated proteins whereas structures with a score higher than 0.5 assume roughly the same fold.A quantitative study
shows that proteins of TM-score = 0.5 have a posterior probability
Posterior probability
In Bayesian statistics, the posterior probability of a random event or an uncertain proposition is the conditional probability that is assigned after the relevant evidence is taken into account...
of 37% in the same CATH
CATH
The CATH Protein Structure Classification is a semi-automatic, hierarchical classification of protein domains published in 1997 by Christine Orengo, Janet Thornton and their colleagues....
Topology family and of 13% in the same SCOP
SCOP
SCOP may refer to:* Structural Classification of Proteins* Suprachiasmatic nucleus circadian oscillatory protein, a member of the leucine-rich repeat protein family* Société coopérative, a type of corporation in France...
Fold family. The probabilities increase rapidly when TM-score >0.5. The TM-score is designed to be independent of protein lengths.
The equation
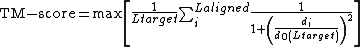
where
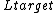 and
and 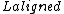 are the lengths of the target protein and the aligned region respectively.
are the lengths of the target protein and the aligned region respectively. 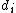 is the distance between the
is the distance between the 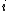 th pair of residues and
th pair of residues and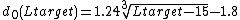
is a distance scale that normalizes distances.
See also
- RMSD — a different structure comparison measure
- GDTGlobal distance testThe global distance test or GDT is a measure of similarity between two protein structures with identical amino acid sequences but different tertiary structures...
— a different structure comparison measure - LCS — a different structure comparison measure
External links
- TM-score webserver — by the Yang Zhang research group. Calculates TM-score and supplies source code.

