
Replisome
Encyclopedia
DNA
Deoxyribonucleic acid is a nucleic acid that contains the genetic instructions used in the development and functioning of all known living organisms . The DNA segments that carry this genetic information are called genes, but other DNA sequences have structural purposes, or are involved in...
. It is made up of a number of subcomponents that each provide a specific function during the process of replication.
Major components
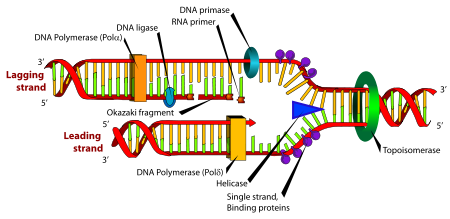
While the replisome acts as a singular machine, its subcomponents are the complexes that perform the specific functions of replication.Helicase
HelicaseHelicase
Helicases are a class of enzymes vital to all living organisms. They are motor proteins that move directionally along a nucleic acid phosphodiester backbone, separating two annealed nucleic acid strands using energy derived from ATP hydrolysis.-Function:Many cellular processes Helicases are a...
is an enzyme which breaks H-bonds between the two strands of DNA. Its doughnut like structure wraps around DNA and separates the strands ahead of DNA synthesis. As helicase unwinds the double helix, it causes the helix to form supercoils in other areas of the DNA. In eukaryotes, the Mcm2-7 complex acts as a helicase, though which subunits are required for helicase activity is not entirely clear. This helicase translocates in the same direction as the DNA polymerase (3' to 5' with respect to the template strand). In prokaryotic systems, the helicases are better identified and include dnaB, which moves 5' to 3' on the strand opposite the DNA polymerase.
Gyrase
Gyrase (a form of Topoisomerase) relaxes and undoes the supercoiling caused by helicase. It does this by cutting the DNA strands, allowing it to rotate and release the supercoil, and then rejoining the strands. Gyrase is most commonly found upstream of the replication fork, where the supercoils form.Primase
PrimasePrimase
DNA primase is an enzyme involved in the replication of DNA.Primase catalyzes the synthesis of a short RNA segment called a primer complementary to a ssDNA template...
adds complementary RNA primers to a DNA strand to begin Okazaki fragments. These primers are approximately 10 base pairs long and are at the 5' end of the okazaki fragment. Additionally, because DNA Polymerase can continue, but not begin, a strand, Primase must begin the leading strand as well.
DNA polymerase III holoenzyme
DNA polymerase III (DNA pol III) holoenzyme contains two catalytic cores - one for replication of the leading strand and one for the lagging strand. DNA pol III stays on the strands via a dimer beta clamp. There is a bridge between the two catalytic cores. DNA pol III also removes the single-strand binding proteins (see below).
DNA pol III has two key limitations:
- It can only add nucleotides to the 3' end of a strand.
- It cannot start a new strand, it can only extend an existing strand (because it must only add to 3' ends of strand).
These limitations directly influence the requirements of the DNA pol III enzyme to both replicated in a leading/lagging fashion, as well as require primers to begin strands.
DNA polymerase I
DNA polymerase IDNA polymerase I
DNA Polymerase I is an enzyme that participates in the process of DNA replication in prokaryotes. It is composed of 928 amino acids, and is an example of a processive enzyme - it can sequentially catalyze multiple polymerisations. Discovered by Arthur Kornberg in 1956, it was the first known...
(DNA pol I) removes the RNA primers set by Primase through exonuclease activity, and (mostly; see ligase below) completes the Okazaki fragments through polymerase activity.
Ligase
Because there is a small gap remaining after pol I continues the strand of the Okazaki fragment, ligaseLigase
In biochemistry, ligase is an enzyme that can catalyse the joining of two large molecules by forming a new chemical bond, usually with accompanying hydrolysis of a small chemical group dependent to one of the larger molecules...
is required to fill in that gap. The two ends of the Okazaki fragments are connected with covalent bonds.
Single-strand binding proteins
Exposed, single-strand DNA is highly unstable (particularly in the aqueous environment of the cell). The single-strand DNA can hydrogen bond to itself and form dangerous hairpin structures. To counteract this instability, single-strand binding proteins (SSB) (e.g. Replication protein AReplication protein A
Replication protein A is a protein that binds single-stranded DNA in eukaryotic cells. During DNA replication, RPA prevents single-stranded DNA from winding back on itself or from forming secondary structures. This keeps DNA unwound for the polymerase to replicate it...
) bind to the exposed bases.

