
Nearest-neighbor chain algorithm
Encyclopedia
In the theory of cluster analysis, the nearest-neighbor chain algorithm is a method that can be used to perform several types of agglomerative hierarchical clustering
, using an amount of memory that is linear in the number of points to be clustered and an amount of time linear in the number of distinct distances between pairs of points. The main idea of the algorithm is to find pairs of clusters to merge by following paths in the nearest neighbor graph
of the clusters until the paths terminate in pairs of mutual nearest neighbors. The algorithm was developed and implemented in 1982 by J. P. Benzécri and J. Juan, based on earlier methods that constructed hierarchical clusterings using mutual nearest neighbor pairs without taking advantage of nearest neighbor chains.
family of clusters with the property that any two clusters in the family are either nested or disjoint.
Alternatively, a hierarchical clustering may be represented as a binary tree
with the points at its leaves; the clusters of the clustering are the sets of points in subtrees descending from each node of the tree.
In agglomerative clustering methods, the input also includes a distance function defined on the points, or a numerical measure of their dissimilarity that is symmetric (insensitive to the ordering within each pair of points) but (unlike a distance) may not satisfy the triangle inequality. Depending on the method, this dissimilarity function can be extended in several different ways to pairs of clusters; for instance, in the single-linkage clustering method, the distance between two clusters is defined to be the minimum distance between any two points from each cluster. Given this distance between clusters, a hierarchical clustering may be defined by a greedy algorithm
that initially places each point in its own single-point cluster and then repeatedly merges the closest pair of clusters.
However, known methods for repeatedly finding the closest pair of clusters in a dynamic set of clusters either require superlinear space to maintain a data structure
that can find closest pairs quickly, or they take greater than linear time to find each closest pair. The nearest-neighbor chain algorithm uses a smaller amount of time and space than the greedy algorithm by merging pairs of clusters in a different order. However, for many types of clustering problem, it can be guaranteed to come up with the same hierarchical clustering as the greedy algorithm despite the different merge order.
of the previous one, until reaching a pair of clusters that are mutual nearest neighbors.
More formally, the algorithm performs the following steps:
If there may be multiple equal nearest neighbors to a cluster, the algorithm requires a consistent tie-breaking rule: for instance, in this case, the nearest neighbor may be chosen, among the clusters at equal minimum distance from , by numbering the clusters arbitrarily and choosing the one with the smallest index.
Since the only data structure is the set of active clusters and the stack containing a subset of the active clusters, the space required is linear in the number of input points.
If a distance function has the reducibility property, then merging two clusters and can only cause the nearest neighbor of to change if that nearest neighbor was one of and . This has two important consequences for the nearest neighbor chain algorithm: first, it can be shown using this property that, at each step of the algorithm, the clusters on the stack form a valid chain of nearest neighbors, because whenever a nearest neighbor becomes invalidated it is immediately removed from the stack.
Second, and even more importantly, it follows from this property that, if two clusters and both belong to the greedy hierarchical clustering, and are mutual nearest neighbors at any point in time, then they will be merged by the greedy clustering, for they must remain mutual nearest neighbors until they are merged. It follows that each mutual nearest neighbor pair found by the nearest neighbor chain algorithm is also a pair of clusters found by the greedy algorithm, and therefore that the nearest neighbor chain algorithm computes exactly the same clustering (although in a different order) as the greedy algorithm.
. That is,
Expressed in terms of the centroid and cardinality
and cardinality 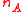 of the two clusters, it has the simpler formula
of the two clusters, it has the simpler formula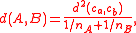
allowing it to be computed in constant time per distance calculation.
Although highly sensitive to outlier
s, Ward's method is the most popular variation of agglomerative clustering both because of the round shape of the clusters it typically forms and because of its principled definition as the clustering that at each step has the smallest variance within its clusters. Alternatively, this distance can be seen as the difference in k-means cost between the new cluster and the two old clusters.
Ward's distance is also reducible, as can be seen more easily from a different formula of Lance–Williams type for calculating the distance of a merged cluster from the distances of the clusters it was merged from: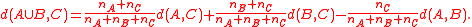
If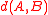 is the smallest of the three distances on the right hand side (as would necessarily be true if
is the smallest of the three distances on the right hand side (as would necessarily be true if  and
and  are mutual nearest-neighbors) then the negative contribution from its term is cancelled by the
are mutual nearest-neighbors) then the negative contribution from its term is cancelled by the 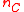 coefficient of one of the two other terms, leaving a positive value added to the weighted average of the other two distances. Therefore, the combined distance is always at least as large as the minimum of
coefficient of one of the two other terms, leaving a positive value added to the weighted average of the other two distances. Therefore, the combined distance is always at least as large as the minimum of 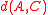 and
and 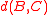 , meeting the definition of reducibility.
, meeting the definition of reducibility.
Therefore, the nearest-neighbor chain algorithm using Ward's distance calculates exactly the same clustering as the standard greedy algorithm. For points in a Euclidean space
of constant dimension, it takes time and space .
or furthest-neighbor clustering is a form of agglomerative clustering that uses the maximum distance between any two points from the two clusters as the dissimilarity, and similarly average-distance clustering uses the average pairwise distance. Like Ward's distance, these forms of clustering obey a formula of Lance-Williams type: in complete linkage, the distance is the average of the distances
is the average of the distances 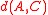 and
and 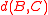 plus a positive correction term, while for average distance it is just a weighted average of the distances
plus a positive correction term, while for average distance it is just a weighted average of the distances 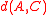 and
and 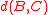 . Thus, in both of these cases, the distance is reducible.
. Thus, in both of these cases, the distance is reducible.
Unlike Ward's method, these two forms of clustering do not have a constant-time method for computing distances between pairs of clusters. Instead it is possible to maintain an array of distances between all pairs of clusters, using the Lance–Williams formula to update the array as pairs of clusters are merged, in time and space . The nearest-neighbor chain algorithm may be used in conjunction with this array of distances to find the same clustering as the greedy algorithm for these cases in total time and space . The same time and space bounds can also be achieved by a different and more general technique that overlays a quadtree
-based priority queue data structure on top of the distance matrix and uses it to perform the standard greedy clustering algorithm, avoiding the need for reducibility, but the nearest-neighbor chain algorithm matches its time and space bounds while using simpler data structures.
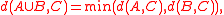
meeting as an equality rather than an inequality the requirement of reducibility. (Single-linkage also obeys a Lance–Williams formula, but with a negative coefficient from which it is more difficult to prove reducibility.)
As with complete linkage and average distance, the difficulty of calculating cluster distances causes the nearest-neighbor chain algorithm to take time and space to compute the single-linkage clustering.
However, the single-linkage clustering can be found more efficiently by an alternative algorithm that computes the minimum spanning tree
of the input distances using Prim's algorithm
(with an unsorted list of vertices and their priorities in place of the usual priority queue), and then sorts the minimum spanning tree edges and uses this sorted list to guide the merger of pairs of clusters. This alternative method would take time and space , matching the best bounds that could be achieved with the nearest-neighbor chain algorithm for distances with constant-time calculations.
to greatly speed up distance computations), care must be taken with recursively defined distances so that they are not using the hierarchy in a way which is sensitive to merge order.
Hierarchical clustering
In statistics, hierarchical clustering is a method of cluster analysis which seeks to build a hierarchy of clusters. Strategies for hierarchical clustering generally fall into two types:...
, using an amount of memory that is linear in the number of points to be clustered and an amount of time linear in the number of distinct distances between pairs of points. The main idea of the algorithm is to find pairs of clusters to merge by following paths in the nearest neighbor graph
Nearest neighbor graph
The nearest neighbor graph for a set of n objects P in a metric space is a directed graph with P being its vertex set and with a directed edge from p to q whenever q is a nearest neighbor of p The nearest neighbor graph (NNG) for a set of n objects P in a metric space (e.g., for a set of points...
of the clusters until the paths terminate in pairs of mutual nearest neighbors. The algorithm was developed and implemented in 1982 by J. P. Benzécri and J. Juan, based on earlier methods that constructed hierarchical clusterings using mutual nearest neighbor pairs without taking advantage of nearest neighbor chains.
Background
The input to a clustering problem consists of a set of points. A cluster is any proper subset of the points, and a hierarchical clustering is a maximalMaximal element
In mathematics, especially in order theory, a maximal element of a subset S of some partially ordered set is an element of S that is not smaller than any other element in S. The term minimal element is defined dually...
family of clusters with the property that any two clusters in the family are either nested or disjoint.
Alternatively, a hierarchical clustering may be represented as a binary tree
Binary tree
In computer science, a binary tree is a tree data structure in which each node has at most two child nodes, usually distinguished as "left" and "right". Nodes with children are parent nodes, and child nodes may contain references to their parents. Outside the tree, there is often a reference to...
with the points at its leaves; the clusters of the clustering are the sets of points in subtrees descending from each node of the tree.
In agglomerative clustering methods, the input also includes a distance function defined on the points, or a numerical measure of their dissimilarity that is symmetric (insensitive to the ordering within each pair of points) but (unlike a distance) may not satisfy the triangle inequality. Depending on the method, this dissimilarity function can be extended in several different ways to pairs of clusters; for instance, in the single-linkage clustering method, the distance between two clusters is defined to be the minimum distance between any two points from each cluster. Given this distance between clusters, a hierarchical clustering may be defined by a greedy algorithm
Greedy algorithm
A greedy algorithm is any algorithm that follows the problem solving heuristic of making the locally optimal choice at each stagewith the hope of finding the global optimum....
that initially places each point in its own single-point cluster and then repeatedly merges the closest pair of clusters.
However, known methods for repeatedly finding the closest pair of clusters in a dynamic set of clusters either require superlinear space to maintain a data structure
Data structure
In computer science, a data structure is a particular way of storing and organizing data in a computer so that it can be used efficiently.Different kinds of data structures are suited to different kinds of applications, and some are highly specialized to specific tasks...
that can find closest pairs quickly, or they take greater than linear time to find each closest pair. The nearest-neighbor chain algorithm uses a smaller amount of time and space than the greedy algorithm by merging pairs of clusters in a different order. However, for many types of clustering problem, it can be guaranteed to come up with the same hierarchical clustering as the greedy algorithm despite the different merge order.
The algorithm
Intuitively, the nearest neighbor chain algorithm repeatedly follows a chain of clusters where each cluster is the nearest neighborNearest neighbor
Nearest neighbor may refer to:* Nearest neighbor search in pattern recognition and in computational geometry* Nearest-neighbor interpolation for interpolating data* Nearest neighbor graph in geometry...
of the previous one, until reaching a pair of clusters that are mutual nearest neighbors.
More formally, the algorithm performs the following steps:
- Initialize the set of active clusters to consist of one-point clusters, one for each input point.
- Let be a stack data structureStack (data structure)In computer science, a stack is a last in, first out abstract data type and linear data structure. A stack can have any abstract data type as an element, but is characterized by only three fundamental operations: push, pop and stack top. The push operation adds a new item to the top of the stack,...
, initially empty, the elements of which will be active clusters. - While there is more than one cluster in the set of clusters:
- If is empty, choose an active cluster arbitrarily and push it onto .
- Let be the active cluster on the top of . Compute the distances from to all the other active clusters, and let be the nearest other active cluster.
- If is already in , it must be the immediate predecessor of . Pop both clusters from , remove them from the set of active clusters, merge the two clusters, and add the merged cluster to the set of active clusters.
- Otherwise, if is not already in , push it onto .
If there may be multiple equal nearest neighbors to a cluster, the algorithm requires a consistent tie-breaking rule: for instance, in this case, the nearest neighbor may be chosen, among the clusters at equal minimum distance from , by numbering the clusters arbitrarily and choosing the one with the smallest index.
Time and space analysis
Each iteration of the loop performs a single search for the nearest neighbor of a cluster, and either adds one cluster to the stack or removes two clusters from it. Every cluster is only ever added once to the stack, because when it is removed again it is immediately made inactive and merged. There are a total of clusters that ever get added to the stack: single-point clusters in the initial set, and internal nodes other than the root in the binary tree representing the clustering. Therefore, the algorithm performs pushing iterations and popping iterations, each time scanning as many as inter-cluster distances to find the nearest neighbor. The total number of distance calculations it makes is therefore less than , and the total time it uses outside of the distance calculations is .Since the only data structure is the set of active clusters and the stack containing a subset of the active clusters, the space required is linear in the number of input points.
Correctness
The correctness of this algorithm relies on a property of its distance function called reducibility, identified by in connection with an earlier clustering method that used mutual nearest neighbor pairs but not chains of nearest neighbors. A distance function on clusters is defined to be reducible if, for every three clusters , and in the greedy hierarchical clustering such that and are mutual nearest neighbors, the following inequality holds:- .
If a distance function has the reducibility property, then merging two clusters and can only cause the nearest neighbor of to change if that nearest neighbor was one of and . This has two important consequences for the nearest neighbor chain algorithm: first, it can be shown using this property that, at each step of the algorithm, the clusters on the stack form a valid chain of nearest neighbors, because whenever a nearest neighbor becomes invalidated it is immediately removed from the stack.
Second, and even more importantly, it follows from this property that, if two clusters and both belong to the greedy hierarchical clustering, and are mutual nearest neighbors at any point in time, then they will be merged by the greedy clustering, for they must remain mutual nearest neighbors until they are merged. It follows that each mutual nearest neighbor pair found by the nearest neighbor chain algorithm is also a pair of clusters found by the greedy algorithm, and therefore that the nearest neighbor chain algorithm computes exactly the same clustering (although in a different order) as the greedy algorithm.
Ward's method
Ward's method is an agglomerative clustering method in which the dissimilarity between two clusters and is measured by the amount by which merging the two clusters into a single larger cluster would increase the average squared distance of a point to its cluster centroidCentroid
In geometry, the centroid, geometric center, or barycenter of a plane figure or two-dimensional shape X is the intersection of all straight lines that divide X into two parts of equal moment about the line. Informally, it is the "average" of all points of X...
. That is,

Expressed in terms of the centroid
 and cardinality
and cardinality 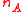 of the two clusters, it has the simpler formula
of the two clusters, it has the simpler formula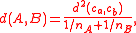
allowing it to be computed in constant time per distance calculation.
Although highly sensitive to outlier
Outlier
In statistics, an outlier is an observation that is numerically distant from the rest of the data. Grubbs defined an outlier as: An outlying observation, or outlier, is one that appears to deviate markedly from other members of the sample in which it occurs....
s, Ward's method is the most popular variation of agglomerative clustering both because of the round shape of the clusters it typically forms and because of its principled definition as the clustering that at each step has the smallest variance within its clusters. Alternatively, this distance can be seen as the difference in k-means cost between the new cluster and the two old clusters.
Ward's distance is also reducible, as can be seen more easily from a different formula of Lance–Williams type for calculating the distance of a merged cluster from the distances of the clusters it was merged from:
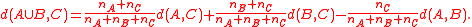
If
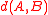 is the smallest of the three distances on the right hand side (as would necessarily be true if
is the smallest of the three distances on the right hand side (as would necessarily be true if  and
and  are mutual nearest-neighbors) then the negative contribution from its term is cancelled by the
are mutual nearest-neighbors) then the negative contribution from its term is cancelled by the 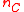 coefficient of one of the two other terms, leaving a positive value added to the weighted average of the other two distances. Therefore, the combined distance is always at least as large as the minimum of
coefficient of one of the two other terms, leaving a positive value added to the weighted average of the other two distances. Therefore, the combined distance is always at least as large as the minimum of 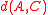 and
and 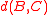 , meeting the definition of reducibility.
, meeting the definition of reducibility.Therefore, the nearest-neighbor chain algorithm using Ward's distance calculates exactly the same clustering as the standard greedy algorithm. For points in a Euclidean space
Euclidean space
In mathematics, Euclidean space is the Euclidean plane and three-dimensional space of Euclidean geometry, as well as the generalizations of these notions to higher dimensions...
of constant dimension, it takes time and space .
Complete linkage and average distance
Complete-linkageComplete-linkage clustering
In cluster analysis, complete linkage or farthest neighbour is a method of calculating distances between clusters in agglomerative hierarchical clustering...
or furthest-neighbor clustering is a form of agglomerative clustering that uses the maximum distance between any two points from the two clusters as the dissimilarity, and similarly average-distance clustering uses the average pairwise distance. Like Ward's distance, these forms of clustering obey a formula of Lance-Williams type: in complete linkage, the distance
 is the average of the distances
is the average of the distances 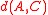 and
and 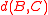 plus a positive correction term, while for average distance it is just a weighted average of the distances
plus a positive correction term, while for average distance it is just a weighted average of the distances 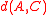 and
and 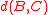 . Thus, in both of these cases, the distance is reducible.
. Thus, in both of these cases, the distance is reducible.Unlike Ward's method, these two forms of clustering do not have a constant-time method for computing distances between pairs of clusters. Instead it is possible to maintain an array of distances between all pairs of clusters, using the Lance–Williams formula to update the array as pairs of clusters are merged, in time and space . The nearest-neighbor chain algorithm may be used in conjunction with this array of distances to find the same clustering as the greedy algorithm for these cases in total time and space . The same time and space bounds can also be achieved by a different and more general technique that overlays a quadtree
Quadtree
A quadtree is a tree data structure in which each internal node has exactly four children. Quadtrees are most often used to partition a two dimensional space by recursively subdividing it into four quadrants or regions. The regions may be square or rectangular, or may have arbitrary shapes. This...
-based priority queue data structure on top of the distance matrix and uses it to perform the standard greedy clustering algorithm, avoiding the need for reducibility, but the nearest-neighbor chain algorithm matches its time and space bounds while using simpler data structures.
Single linkage
In single-linkage or nearest-neighbor clustering, the oldest form of agglomerative hierarchical clustering, the dissimilarity between clusters is measured as the minimum distance between any two points from the two clusters. With this dissimilarity,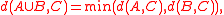
meeting as an equality rather than an inequality the requirement of reducibility. (Single-linkage also obeys a Lance–Williams formula, but with a negative coefficient from which it is more difficult to prove reducibility.)
As with complete linkage and average distance, the difficulty of calculating cluster distances causes the nearest-neighbor chain algorithm to take time and space to compute the single-linkage clustering.
However, the single-linkage clustering can be found more efficiently by an alternative algorithm that computes the minimum spanning tree
Minimum spanning tree
Given a connected, undirected graph, a spanning tree of that graph is a subgraph that is a tree and connects all the vertices together. A single graph can have many different spanning trees...
of the input distances using Prim's algorithm
Prim's algorithm
In computer science, Prim's algorithm is a greedy algorithm that finds a minimum spanning tree for a connected weighted undirected graph. This means it finds a subset of the edges that forms a tree that includes every vertex, where the total weight of all the edges in the tree is minimized...
(with an unsorted list of vertices and their priorities in place of the usual priority queue), and then sorts the minimum spanning tree edges and uses this sorted list to guide the merger of pairs of clusters. This alternative method would take time and space , matching the best bounds that could be achieved with the nearest-neighbor chain algorithm for distances with constant-time calculations.
Centroid distance
Another distance measure commonly used in agglomerative clustering is the distance between the centroids of pairs of clusters, also known as the weighted group method. It can be calculated easily in constant time per distance calculation. However, it is not reducible: for instance, if the input forms the set of three points of an equilateral triangle, merging two of these points into a larger cluster causes the inter-cluster distance to decrease, a violation of reducibility. Therefore, the nearest-neighbor chain algorithm will not necessarily find the same clustering as the greedy algorithm. A different algorithm by Day and Edelsbrunner can be used to find the clustering in time for this distance measure.Distances sensitive to merge order
The above presentation explicitly disallowed distances sensitive to merge order; indeed, allowing such distances can cause problems. In particular, there exist order-sensitive cluster distances which satisfy reducibility, but the above algorithm will return a hierarchy with suboptimal costs. Following the earlier discussion of the value of defining cluster distances recursively (so that memoization can be usedto greatly speed up distance computations), care must be taken with recursively defined distances so that they are not using the hierarchy in a way which is sensitive to merge order.

