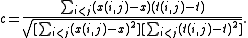Cophenetic correlation
Encyclopedia
In statistics
, and especially in biostatistics
, cophenetic correlation (more precisely, the cophenetic correlation coefficient) is a measure of how faithfully a dendrogram
preserves the pairwise distances between the original unmodeled data points. Although it has been most widely applied in the field of biostatistics (typically to assess cluster-based models of DNA
sequences, or other taxonomic models), it can also be used in other fields of inquiry where raw data tend to occur in clumps, or clusters. This coefficient has also been proposed for use as a test for nested clusters.
Then, letting x be the average of the x(i, j), and letting t be the average of the t(i, j), the cophenetic correlation coefficient c is given by
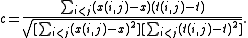
Statistics
Statistics is the study of the collection, organization, analysis, and interpretation of data. It deals with all aspects of this, including the planning of data collection in terms of the design of surveys and experiments....
, and especially in biostatistics
Biostatistics
Biostatistics is the application of statistics to a wide range of topics in biology...
, cophenetic correlation (more precisely, the cophenetic correlation coefficient) is a measure of how faithfully a dendrogram
Dendrogram
A dendrogram is a tree diagram frequently used to illustrate the arrangement of the clusters produced by hierarchical clustering...
preserves the pairwise distances between the original unmodeled data points. Although it has been most widely applied in the field of biostatistics (typically to assess cluster-based models of DNA
DNA
Deoxyribonucleic acid is a nucleic acid that contains the genetic instructions used in the development and functioning of all known living organisms . The DNA segments that carry this genetic information are called genes, but other DNA sequences have structural purposes, or are involved in...
sequences, or other taxonomic models), it can also be used in other fields of inquiry where raw data tend to occur in clumps, or clusters. This coefficient has also been proposed for use as a test for nested clusters.
Calculating the cophenetic correlation coefficient
Suppose that the original data {Xi} have been modeled using a cluster method to produce a dendrogram {Ti}; that is, a simplified model in which data that are "close" have been grouped into a hierarchical tree. Define the following distance measures.- x(i, j) = | Xi − Xj |, the ordinary Euclidean distance between the ith and jth observations.
- t(i, j) = the dendrogrammatic distance between the model points Ti and Tj. This distance is the height of the node at which these two points are first joined together.
Then, letting x be the average of the x(i, j), and letting t be the average of the t(i, j), the cophenetic correlation coefficient c is given by