
A-DNA
Encyclopedia
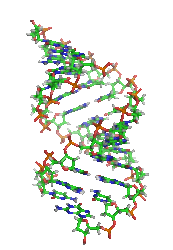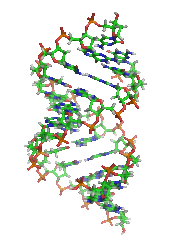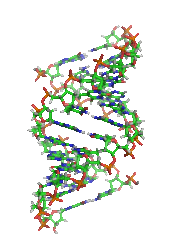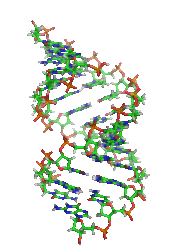
DNA
Deoxyribonucleic acid is a nucleic acid that contains the genetic instructions used in the development and functioning of all known living organisms . The DNA segments that carry this genetic information are called genes, but other DNA sequences have structural purposes, or are involved in...
. A-DNA is thought to be one of three biologically active double helical structures along with B- and Z-DNA
Z-DNA
Z-DNA is one of the many possible double helical structures of DNA. It is a left-handed double helical structure in which the double helix winds to the left in a zig-zag pattern...
. It is a right-handed double helix fairly similar to the more common and well-known B-DNA form, but with a shorter more compact helical structure. It appears likely that it occurs only in dehydrated samples of DNA, such as those used in crystallographic experiments, and possibly is also assumed by DNA-RNA hybrid helices and by regions of double-stranded RNA.
Structure
A-DNA is fairly similar to B-DNA given that it is a right-handed double helix with major and minor grooves. However, as shown in the comparison table below, there is a slight increase in the number of base pairs per rotation (resulting in a tighter rotation angle), and smaller rise/turn. This results in a deepening of the major groove and a shallowing of the minor.Predicting A-DNA structure
An algorithm for predicting the propensity of a sequence to flip from B-DNA to A-DNA was developed by Beth Basham, Gary Schroth, and P. Shing Ho at Oregon State UniversityOregon State University
Oregon State University is a coeducational, public research university located in Corvallis, Oregon, United States. The university offers undergraduate, graduate and doctoral degrees and a multitude of research opportunities. There are more than 200 academic degree programs offered through the...
.
The abstract of their work describes this algorithm:
The ability to predict macromolecular conformations from sequence and thermodynamic principles has long been coveted but generally has not been achieved. We show that differences in the hydration of DNA surfaces can be used to distinguish between sequences that form A- and B-DNA. From this, a "triplet code" of A-DNA propensities was derived as energetic rules for predicting A-DNA formation. This code correctly predicted > 90% of A- and B-DNA sequences in crystals and correlates with A-DNA formation in solution. Thus, with our previous studies on Z-DNA, we now have a single method to predict the relative stability of sequences in the three standard DNA duplex conformations.
Comparison Geometries of the Most Common DNA Forms


| Geometry attribute: | A-form | B-form | Z-form |
|---|---|---|---|
| Helix sense | right-handed | right-handed | left-handed |
| Repeating unit | 1 bp | 1 bp | 2 bp |
| Rotation/bp | 33.6° | 35.9° | 60°/2 |
| Mean bp/turn | 11 | 10.5 | 12 |
| Inclination of bp to axis | +19° | −1.2° | −9° |
| Rise/bp along axis | 2.4 Å (0.24 nm) | 3.4 Å (0.34 nm) | 3.7 Å (0.37 nm) |
| Rise/turn of helix | 24.6 Å (2.46 nm) | 33.2 Å (3.32 nm) | 45.6 Å (4.56 nm) |
| Mean propeller twist | +18° | +16° | 0° |
| Glycosyl angle | anti | anti | pyrimidine: anti, purine: syn |
| Sugar pucker | C3'-endo | C2'-endo | C: C2'-endo, G: C2'-exo |
| Diameter | 23 Å (2.3 nm) | 20 Å (2.0 nm) | 18 Å (1.8 nm) |

