
Wagner's gene network model
Encyclopedia
Wagner's gene network model is a computational model of artificial gene networks, which explicitly modeled the developmental and evolutionary process of genetic regulatory networks. A population
with multiple organisms can be created and evolved from generation to generation. It was first developed by Andreas Wagner in 1996 and has been investigated by other groups to study the evolution of gene networks, gene expression
, robustness
, plasticity
and epistasis
.
 genes encoding transcription factors. The product of each gene can regulate the expression level of itself and/or the other genes through cis-regulatory element
genes encoding transcription factors. The product of each gene can regulate the expression level of itself and/or the other genes through cis-regulatory element
s. The interactions among genes constitute a gene network that is represented by a ×
×  regulatory matrix
regulatory matrix 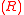 in the model. The elements in matrix R represent the interaction strength. Positive values within the matrix represent the activation of the target gene, while negative ones represent repression. Matrix elements with value 0 indicate the absence of interactions between two genes.
in the model. The elements in matrix R represent the interaction strength. Positive values within the matrix represent the activation of the target gene, while negative ones represent repression. Matrix elements with value 0 indicate the absence of interactions between two genes.
pattern at time . It is represented by a state vector
. It is represented by a state vector  in this model.
in this model.
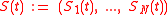
whose elements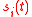 denotes the expression states of gene i at time t. In the original Wagner model,
denotes the expression states of gene i at time t. In the original Wagner model,
 ∈
∈ 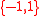
where 1 represents the gene is expressed while -1 implies the gene is not expressed. The expression pattern can only be ON or OFF. The continuous expression pattern between -1 (or 0) and 1 is also implemented in some other variants.
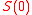 at time
at time  is defined as the initial expression state. The interactions among genes change the expression states during the development process. This process is modeled by the following differential equations
is defined as the initial expression state. The interactions among genes change the expression states during the development process. This process is modeled by the following differential equations
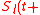 τ
τ σ
σ 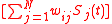
σ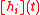
where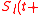 τ) represents the expression state of
τ) represents the expression state of  at time t+τ. It is determined by a filter function σ
at time t+τ. It is determined by a filter function σ .
.  represents the weighted sum of regulatory effects (
represents the weighted sum of regulatory effects (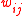 ) of all genes on gene
) of all genes on gene  at time t. In the original Wagner model, the filter function is a step function
at time t. In the original Wagner model, the filter function is a step function
σ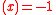 if
if  if
if  if
if 
In other variants, the filter function is implemented as a sigmoidal function
σ
In this way, the expression states will acquire a continuous distribution. The gene expression will reach the final state if it reaches a stable pattern.
 .
.
and asexual reproduction
s are implemented. Asexual reproduction is implemented as producing the offspring's genome
(the gene network) by directly copying the parent's genome. Sexual reproduction is implemented as the recombination of the two parents' genomes.
Population
A population is all the organisms that both belong to the same group or species and live in the same geographical area. The area that is used to define a sexual population is such that inter-breeding is possible between any pair within the area and more probable than cross-breeding with individuals...
with multiple organisms can be created and evolved from generation to generation. It was first developed by Andreas Wagner in 1996 and has been investigated by other groups to study the evolution of gene networks, gene expression
Gene expression
Gene expression is the process by which information from a gene is used in the synthesis of a functional gene product. These products are often proteins, but in non-protein coding genes such as ribosomal RNA , transfer RNA or small nuclear RNA genes, the product is a functional RNA...
, robustness
Mutational robustness
Mutational robustness describes the extent to which an organism’s phenotype remains constant in spite of mutation. Natural selection can directly induce the evolution of mutational robustness only when mutation rates are high and population sizes are large...
, plasticity
Phenotypic plasticity
Phenotypic plasticity is the ability of an organism to change its phenotype in response to changes in the environment. Such plasticity in some cases expresses as several highly morphologically distinct results; in other cases, a continuous norm of reaction describes the functional interrelationship...
and epistasis
Epistasis
In genetics, epistasis is the phenomenon where the effects of one gene are modified by one or several other genes, which are sometimes called modifier genes. The gene whose phenotype is expressed is called epistatic, while the phenotype altered or suppressed is called hypostatic...
.
Assumptions
The model and its variants have a number of simplifying assumptions. Three of them are listing below.- The organisms are modeled as gene regulatory networks. The models assume that gene expression is regulated exclusively at the transcriptional level;
- The product of a gene can regulate the expression of (be a regulator of) that source gene or other genes. The models assume that a gene can only produce one active transcriptional regulator;
- The effects of one regulator are independent of effects of other regulators on the same target gene.
Genotype
The model represents individuals as networks of interacting transcriptional regulators. Each individual expresses genes encoding transcription factors. The product of each gene can regulate the expression level of itself and/or the other genes through cis-regulatory element
genes encoding transcription factors. The product of each gene can regulate the expression level of itself and/or the other genes through cis-regulatory elementCis-regulatory element
A cis-regulatory element or cis-element is a region of DNA or RNA that regulates the expression of genes located on that same molecule of DNA . This term is constructed from the Latin word cis, which means "on the same side as". These cis-regulatory elements are often binding sites for one or...
s. The interactions among genes constitute a gene network that is represented by a
 ×
×  regulatory matrix
regulatory matrix 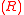 in the model. The elements in matrix R represent the interaction strength. Positive values within the matrix represent the activation of the target gene, while negative ones represent repression. Matrix elements with value 0 indicate the absence of interactions between two genes.
in the model. The elements in matrix R represent the interaction strength. Positive values within the matrix represent the activation of the target gene, while negative ones represent repression. Matrix elements with value 0 indicate the absence of interactions between two genes.Phenotype
The phenotype of each individual is modeled as the gene expressionGene expression
Gene expression is the process by which information from a gene is used in the synthesis of a functional gene product. These products are often proteins, but in non-protein coding genes such as ribosomal RNA , transfer RNA or small nuclear RNA genes, the product is a functional RNA...
pattern at time
 . It is represented by a state vector
. It is represented by a state vector  in this model.
in this model.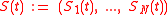
whose elements
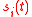 denotes the expression states of gene i at time t. In the original Wagner model,
denotes the expression states of gene i at time t. In the original Wagner model, ∈
∈ 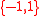
where 1 represents the gene is expressed while -1 implies the gene is not expressed. The expression pattern can only be ON or OFF. The continuous expression pattern between -1 (or 0) and 1 is also implemented in some other variants.
Development
The development process is modeled as the development of gene expression states. The gene expression pattern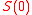 at time
at time  is defined as the initial expression state. The interactions among genes change the expression states during the development process. This process is modeled by the following differential equations
is defined as the initial expression state. The interactions among genes change the expression states during the development process. This process is modeled by the following differential equations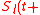 τ
τ σ
σ 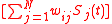
σ
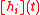
where
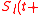 τ) represents the expression state of
τ) represents the expression state of  at time t+τ. It is determined by a filter function σ
at time t+τ. It is determined by a filter function σ .
.  represents the weighted sum of regulatory effects (
represents the weighted sum of regulatory effects (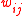 ) of all genes on gene
) of all genes on gene  at time t. In the original Wagner model, the filter function is a step function
at time t. In the original Wagner model, the filter function is a step functionσ
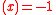 if
if  if
if  if
if 
In other variants, the filter function is implemented as a sigmoidal function
σ

In this way, the expression states will acquire a continuous distribution. The gene expression will reach the final state if it reaches a stable pattern.
Evolution
Evolutionary simulations are performed by reproduction-mutation-selection life cycle. Populations are fixed at size N and they will not go extinct. Non-overlapping generations are employed. In a typical evolutionary simulation, a single random viable individual that can produce a stable gene expression pattern is chosen as the founder. Cloned individuals are generated to create a population of N identical individuals. According to the asexual or sexual reproductive mode, offsprings are produced by randomly choosing (with replacement) parent individual(s) from current generation. Mutations can be acquired with probability μ and survive with probability equal to their fitness. This process is repeated until N individuals are produced that go on to found the following generation.Fitness
Fitness in this model is the probability that an individual survives to reproduce. In the simplest implementation of the model, developmentally stable genotypes survive (i.e. their fitness is 1) and develop- mentally unstable ones do not (i.e. their fitness is 0).Mutation
Mutations are modeled as the changes in gene regulation, i.e., the changes of the elements in the regulatory matrix .
.Reproduction
Both sexualSexual reproduction
Sexual reproduction is the creation of a new organism by combining the genetic material of two organisms. There are two main processes during sexual reproduction; they are: meiosis, involving the halving of the number of chromosomes; and fertilization, involving the fusion of two gametes and the...
and asexual reproduction
Asexual reproduction
Asexual reproduction is a mode of reproduction by which offspring arise from a single parent, and inherit the genes of that parent only, it is reproduction which does not involve meiosis, ploidy reduction, or fertilization. A more stringent definition is agamogenesis which is reproduction without...
s are implemented. Asexual reproduction is implemented as producing the offspring's genome
Genome
In modern molecular biology and genetics, the genome is the entirety of an organism's hereditary information. It is encoded either in DNA or, for many types of virus, in RNA. The genome includes both the genes and the non-coding sequences of the DNA/RNA....
(the gene network) by directly copying the parent's genome. Sexual reproduction is implemented as the recombination of the two parents' genomes.

